1GQ2
 
 | | Malic Enzyme from Pigeon Liver | | Descriptor: | CHLORIDE ION, MALIC ENZYME, MANGANESE (II) ION, ... | | Authors: | Yang, Z, Zhang, H, Liang, T. | | Deposit date: | 2001-11-19 | | Release date: | 2002-05-23 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural Studies of the Pigeon Cytosolic Nadp+ -Dependent Malic Enzyme
Protein Sci., 11, 2002
|
|
3IT8
 
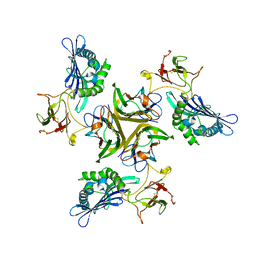 | |
1Q0S
 
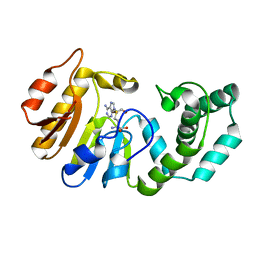 | | Binary Structure of T4DAM with AdoHcy | | Descriptor: | DNA adenine methylase, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Yang, Z, Horton, J.R, Zhou, L, Zhang, X.J, Dong, A, Zhang, X, Schlagman, S.L, Kossykh, V, Hattman, S, Cheng, X. | | Deposit date: | 2003-07-17 | | Release date: | 2003-09-23 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of the bacteriophage T4 DNA adenine methyltransferase
Nat.Struct.Biol., 10, 2003
|
|
3D2U
 
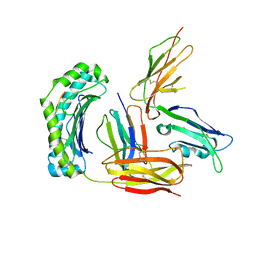 | | Structure of UL18, a Peptide-Binding Viral MHC Mimic, Bound to a Host Inhibitory Receptor | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Actin, Beta-2-microglobulin, ... | | Authors: | Yang, Z, Bjorkman, P.J. | | Deposit date: | 2008-05-08 | | Release date: | 2008-07-08 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Structure of UL18, a peptide-binding viral MHC mimic, bound to a host inhibitory receptor
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
1Q0T
 
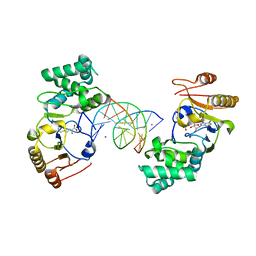 | | Ternary Structure of T4DAM with AdoHcy and DNA | | Descriptor: | 5'-D(*AP*CP*AP*GP*GP*AP*TP*CP*CP*TP*GP*T)-3', DNA adenine methylase, IODIDE ION, ... | | Authors: | Yang, Z, Horton, J.R, Zhou, L, Zhang, X.J, Dong, A, Zhang, X, Schlagman, S.L, Kossykh, V, Hattman, S, Cheng, X. | | Deposit date: | 2003-07-17 | | Release date: | 2003-09-23 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structure of the bacteriophage T4 DNA adenine methyltransferase
Nat.Struct.Biol., 10, 2003
|
|
1PJL
 
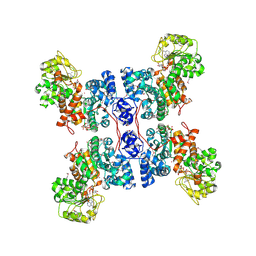 | | Crystal structure of human m-NAD-ME in ternary complex with NAD and Lu3+ | | Descriptor: | LUTETIUM (III) ION, NAD-dependent malic enzyme, mitochondrial, ... | | Authors: | Yang, Z, Batra, R, Floyd, D.L, Hung, H.-C, Chang, G.-G, Tong, L. | | Deposit date: | 2003-06-03 | | Release date: | 2003-06-17 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Potent and competitive inhibition of malic enzymes by lanthanide ions
Biochem.Biophys.Res.Commun., 274, 2000
|
|
1T43
 
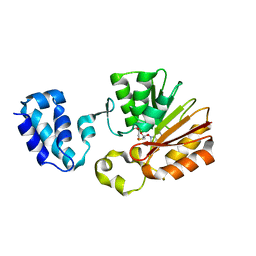 | | Crystal Structure Analysis of E.coli Protein (N5)-Glutamine Methyltransferase (HemK) | | Descriptor: | Protein methyltransferase hemK, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Yang, Z, Shipman, L, Zhang, M, Anton, B.P, Roberts, R.J, Cheng, X. | | Deposit date: | 2004-04-28 | | Release date: | 2004-06-29 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural characterization and comparative phylogenetic analysis of Escherichia coli HemK, a protein (N5)-glutamine methyltransferase.
J.Mol.Biol., 340, 2004
|
|
1YNM
 
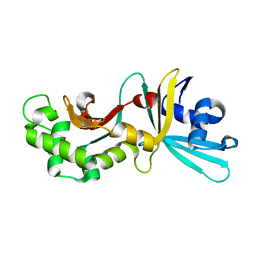 | | Crystal structure of restriction endonuclease HinP1I | | Descriptor: | R.HinP1I restriction endonuclease | | Authors: | Yang, Z, Horton, J.R, Maunus, R, Wilson, G.G, Roberts, R.J, Cheng, X. | | Deposit date: | 2005-01-24 | | Release date: | 2005-05-03 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structure of HinP1I endonuclease reveals a striking similarity to the monomeric restriction enzyme MspI
Nucleic Acids Res., 33, 2005
|
|
6U0N
 
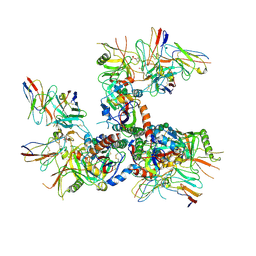 | |
6U0L
 
 | |
7RYV
 
 | |
7RYU
 
 | |
8D5C
 
 | | anti-HIV-1 gp120-sCD4 complex antibody CG10 Fab in complex with B41-sCD4 | | Descriptor: | CG10 Fab heavy chain, CG10 Fab light chain, Envelope glycoprotein gp160, ... | | Authors: | Yang, Z, Bjorkman, P.J, Gershoni, J.M. | | Deposit date: | 2022-06-04 | | Release date: | 2022-11-16 | | Last modified: | 2023-01-04 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Antibody Recognition of CD4-Induced Open HIV-1 Env Trimers.
J.Virol., 96, 2022
|
|
8D54
 
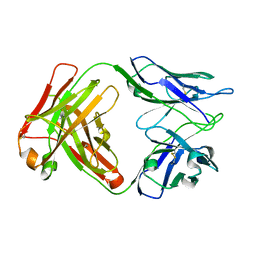 | |
2LUH
 
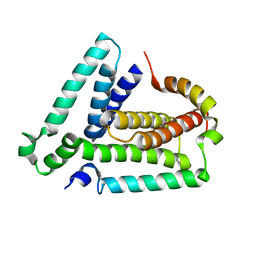 | | NMR structure of the Vta1-Vps60 complex | | Descriptor: | Vacuolar protein sorting-associated protein VTA1, Vacuolar protein-sorting-associated protein 60 | | Authors: | Yang, Z, Vild, C, Ju, J, Zhang, X, Liu, J, Shen, J, Zhao, B, Lan, W, Gong, F, Liu, M, Cao, C, Xu, Z. | | Deposit date: | 2012-06-13 | | Release date: | 2012-11-07 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural Basis of Molecular Recognition between ESCRT-III-like Protein Vps60 and AAA-ATPase Regulator Vta1 in the Multivesicular Body Pathway.
J.Biol.Chem., 287, 2012
|
|
1EFL
 
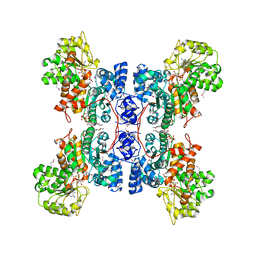 | | HUMAN MALIC ENZYME IN A QUATERNARY COMPLEX WITH NAD, MG, AND TARTRONATE | | Descriptor: | MAGNESIUM ION, MALIC ENZYME, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Yang, Z, Floyd, D.L, Loeber, G, Tong, L. | | Deposit date: | 2000-02-09 | | Release date: | 2000-03-08 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of a closed form of human malic enzyme and implications for catalytic mechanism.
Nat.Struct.Biol., 7, 2000
|
|
1DO8
 
 | | CRYSTAL STRUCTURE OF A CLOSED FORM OF HUMAN MITOCHONDRIAL NAD(P)+-DEPENDENT MALIC ENZYME | | Descriptor: | MALIC ENZYME, MANGANESE (II) ION, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Yang, Z, Floyd, D.L, Loeber, G, Tong, L. | | Deposit date: | 1999-12-19 | | Release date: | 2000-03-06 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of a closed form of human malic enzyme and implications for catalytic mechanism.
Nat.Struct.Biol., 7, 2000
|
|
1EI3
 
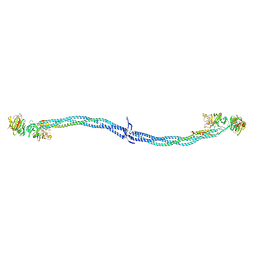 | | CRYSTAL STRUCTURE OF NATIVE CHICKEN FIBRINOGEN | | Descriptor: | FIBRINOGEN | | Authors: | Yang, Z, Mochalkin, I, Veerapandian, L, Riley, M, Doolittle, R.F. | | Deposit date: | 2000-02-23 | | Release date: | 2000-05-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (5.5 Å) | | Cite: | Crystal structure of native chicken fibrinogen at 5.5-A resolution.
Proc.Natl.Acad.Sci.USA, 97, 2000
|
|
1EFK
 
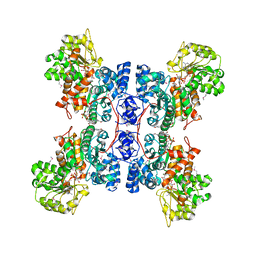 | | STRUCTURE OF HUMAN MALIC ENZYME IN COMPLEX WITH KETOMALONATE | | Descriptor: | ALPHA-KETOMALONIC ACID, MAGNESIUM ION, MALIC ENZYME, ... | | Authors: | Yang, Z, Floyd, D.L, Loeber, G, Tong, L. | | Deposit date: | 2000-02-09 | | Release date: | 2000-03-08 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of a closed form of human malic enzyme and implications for catalytic mechanism.
Nat.Struct.Biol., 7, 2000
|
|
7RW4
 
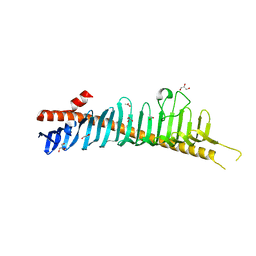 | | Crystal structure of junctophilin-1 | | Descriptor: | ACETATE ION, GLYCEROL, Junctophilin-1 | | Authors: | Yang, Z, Panwar, P, Van Petegem, F. | | Deposit date: | 2021-08-19 | | Release date: | 2022-02-23 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.31 Å) | | Cite: | Structures of the junctophilin/voltage-gated calcium channel interface reveal hot spot for cardiomyopathy mutations.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7RXE
 
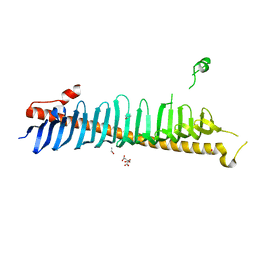 | | Crystal structure of junctophilin-2 | | Descriptor: | CITRATE ANION, ISOPROPYL ALCOHOL, Junctophilin-2 N-terminal fragment | | Authors: | Yang, Z, Panwar, P, Van Petegem, F. | | Deposit date: | 2021-08-22 | | Release date: | 2022-02-23 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structures of the junctophilin/voltage-gated calcium channel interface reveal hot spot for cardiomyopathy mutations.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7RXQ
 
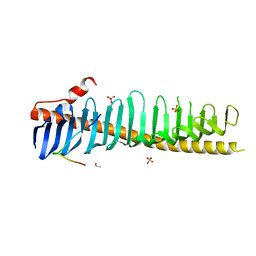 | | Crystal structure of junctophilin-2 in complex with a CaV1.1 peptide | | Descriptor: | ETHANOL, Junctophilin-2 N-terminal fragment, SULFATE ION, ... | | Authors: | Yang, Z, Panwar, P, Van Petegem, F. | | Deposit date: | 2021-08-23 | | Release date: | 2022-02-23 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Structures of the junctophilin/voltage-gated calcium channel interface reveal hot spot for cardiomyopathy mutations.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
3K6J
 
 | | Crystal structure of the dehydrogenase part of multifuctional enzyme 1 from C.elegans | | Descriptor: | PHOSPHATE ION, Protein F01G10.3, confirmed by transcript evidence, ... | | Authors: | Ouyang, Z, Zhang, K, Zhai, Y, Lu, J, Sun, F. | | Deposit date: | 2009-10-09 | | Release date: | 2010-10-13 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of the dehydrogenase part of multifuctional enzyme 1 from C.elegans
To be Published
|
|
5FIS
 
 | | Exonuclease domain-containing 1 (Exd1) in the Gd bound conformation | | Descriptor: | EXD1, GADOLINIUM ATOM | | Authors: | Yang, Z, Chen, K.M, Pandey, R.R, Homolka, D, Reuter, M, Rodino Janeiro, B.K, Sachidanandam, R, Fauvarque, M.O, McCarthy, A.A, Pillai, R.S. | | Deposit date: | 2015-10-02 | | Release date: | 2015-12-23 | | Last modified: | 2016-01-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Piwi Slicing and Exd1 Drive Biogenesis of Nuclear Pirnas from Cytosolic Targets of the Mouse Pirna Pathway
Mol.Cell, 61, 2016
|
|
5FIQ
 
 | | Exonuclease domain-containing 1 (Exd1) in the native conformation | | Descriptor: | EXD1 | | Authors: | Yang, Z, Chen, K.M, Pandey, R.R, Homolka, D, Reuter, M, Rodino Janeiro, B.K, Sachidanandam, R, Fauvarque, M.O, McCarthy, A.A, Pillai, R.S. | | Deposit date: | 2015-10-01 | | Release date: | 2015-12-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Piwi Slicing and Exd1 Drive Biogenesis of Nuclear Pirnas from Cytosolic Targets of the Mouse Pirna Pathway
Mol.Cell, 61, 2016
|
|
