6KCZ
 
 | |
4M02
 
 | | Middle fragment(residues 494-663) of the binding region of SraP | | Descriptor: | CALCIUM ION, GLYCEROL, Serine-rich adhesin for platelets | | Authors: | Yang, Y.H, Jiang, Y.L, Zhang, J, Wang, L, Chen, Y, Zhou, C.Z. | | Deposit date: | 2013-08-01 | | Release date: | 2014-06-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Structural Insights into SraP-Mediated Staphylococcus aureus Adhesion to Host Cells
Plos Pathog., 10, 2014
|
|
4M03
 
 | | C-terminal fragment(residues 576-751) of binding region of SraP | | Descriptor: | CALCIUM ION, Serine-rich adhesin for platelets | | Authors: | Yang, Y.H, Jiang, Y.L, Zhang, J, Wang, L, Chen, Y, Zhou, C.Z. | | Deposit date: | 2013-08-01 | | Release date: | 2014-06-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Structural Insights into SraP-Mediated Staphylococcus aureus Adhesion to Host Cells
Plos Pathog., 10, 2014
|
|
4M01
 
 | | N terminal fragment(residues 245-575) of binding region of SraP | | Descriptor: | CALCIUM ION, GLYCEROL, Serine-rich adhesin for platelets | | Authors: | Yang, Y.H, Jiang, Y.L, Zhang, J, Wang, L, Chen, Y, Zhou, C.Z. | | Deposit date: | 2013-08-01 | | Release date: | 2014-06-18 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural Insights into SraP-Mediated Staphylococcus aureus Adhesion to Host Cells
Plos Pathog., 10, 2014
|
|
5ZZ3
 
 | | Crystal structure of intracellular B30.2 domain of BTN3A3 | | Descriptor: | Butyrophilin, subfamily 3, member A3 isoform b variant | | Authors: | Yang, Y.Y, Li, X, Liu, W.D, Chen, C.C, Guo, R.T, Zhang, Y.H. | | Deposit date: | 2018-05-30 | | Release date: | 2019-04-03 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | A Structural Change in Butyrophilin upon Phosphoantigen Binding Underlies Phosphoantigen-Mediated V gamma 9V delta 2 T Cell Activation.
Immunity, 50, 2019
|
|
5ZXK
 
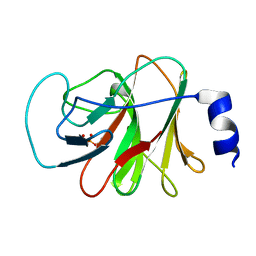 | | Crystal structure of intracellular B30.2 domain of BTN3A1 in complex with HMBPP | | Descriptor: | (2E)-4-hydroxy-3-methylbut-2-en-1-yl trihydrogen diphosphate, Butyrophilin subfamily 3 member A1 | | Authors: | Yang, Y.Y, Liu, W.D, Chen, C.C, Guo, R.T, Zhang, Y.H. | | Deposit date: | 2018-05-21 | | Release date: | 2019-04-03 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | A Structural Change in Butyrophilin upon Phosphoantigen Binding Underlies Phosphoantigen-Mediated V gamma 9V delta 2 T Cell Activation.
Immunity, 50, 2019
|
|
4M00
 
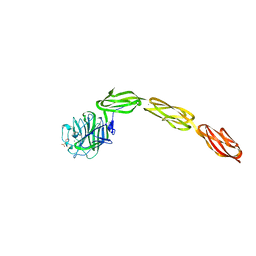 | | Crystal structure of the ligand binding region of staphylococcal adhesion SraP | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, Serine-rich adhesin for platelets, ... | | Authors: | Yang, Y.H, Jiang, Y.L, Zhang, J, Wang, L, Chen, Y, Zhou, C.Z. | | Deposit date: | 2013-08-01 | | Release date: | 2014-06-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural Insights into SraP-Mediated Staphylococcus aureus Adhesion to Host Cells
Plos Pathog., 10, 2014
|
|
6KZE
 
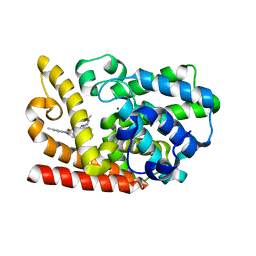 | | The crystal structue of PDE10A complexed with 4d | | Descriptor: | 8-[(E)-2-[5-methyl-1-[3-[3-(4-methylpiperazin-1-yl)propoxy]phenyl]benzimidazol-2-yl]ethenyl]quinoline, MAGNESIUM ION, ZINC ION, ... | | Authors: | Yang, Y, Zhang, S, Zhou, Q, Huang, Y.-Y, Guo, L, Luo, H.-B. | | Deposit date: | 2019-09-24 | | Release date: | 2020-09-30 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.50003481 Å) | | Cite: | Novel Potent and Highly Selective Benzoimidazole-based Phosphodiesterase 10 Inhibitors with Improved Solubility and Pharmacokinetic Properties for the Treatment of Pulmonary Arterial Hypertension
To Be Published
|
|
7E3U
 
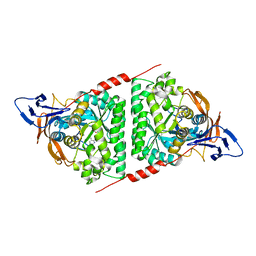 | | Crystal structure of the Pseudomonas aeruginosa dihydropyrimidinase complexed with 5-AU | | Descriptor: | 5-AMINO-1H-PYRIMIDINE-2,4-DIONE, D-hydantoinase/dihydropyrimidinase, ZINC ION | | Authors: | Yang, Y.C, Luo, R.H, Huang, Y.H, Huang, C.Y, Lin, E.S. | | Deposit date: | 2021-02-09 | | Release date: | 2022-02-16 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.159 Å) | | Cite: | Molecular Insights into How the Dimetal Center in Dihydropyrimidinase Can Bind the Thymine Antagonist 5-Aminouracil: A Different Binding Mode from the Anticancer Drug 5-Fluorouracil.
Bioinorg Chem Appl, 2022, 2022
|
|
7EYI
 
 | |
7DUU
 
 | | Crystal structure of HLA molecule with an KIR receptor | | Descriptor: | Beta-2-microglobulin, Killer cell immunoglobulin-like receptor 2DS2, LEU-ASN-PRO-SER-VAL-ALA-ALA-THR-LEU, ... | | Authors: | Yang, Y, Yin, L. | | Deposit date: | 2021-01-11 | | Release date: | 2022-02-02 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Activating receptor KIR2DS2 bound to HLA-C1 reveals the novel recognition features of activating receptor.
Immunology, 165, 2022
|
|
7CYW
 
 | |
4XYZ
 
 | | Crystal structure of K33 linked di-Ubiquitin | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, IODIDE ION, ... | | Authors: | Kristariyanto, Y.A, Abdul Rehman, S.A, Choi, S.Y, Ritorto, S, Campbell, D.G, Morrice, N.A, Toth, R, Kulathu, Y. | | Deposit date: | 2015-02-03 | | Release date: | 2015-03-18 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Assembly and structure of Lys33-linked polyubiquitin reveals distinct conformations.
Biochem.J., 467, 2015
|
|
4Y1H
 
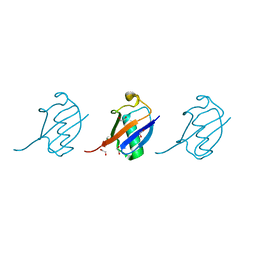 | | Crystal structure of K33 linked tri-Ubiquitin | | Descriptor: | 1,2-ETHANEDIOL, Ubiquitin-40S ribosomal protein S27a | | Authors: | Kristariyanto, Y.A, Abdul Rehman, S.A, Choi, S.Y, Ritorto, S, Campbell, D.G, Morrice, N.A, Toth, R, Kulathu, Y. | | Deposit date: | 2015-02-07 | | Release date: | 2015-03-18 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Assembly and structure of Lys33-linked polyubiquitin reveals distinct conformations.
Biochem.J., 467, 2015
|
|
9CX6
 
 | |
7Q4M
 
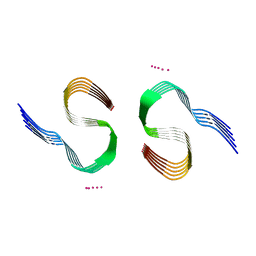 | | Type II beta-amyloid 42 Filaments from Human Brain | | Descriptor: | Amyloid-beta precursor protein, UNKNOWN ATOM OR ION | | Authors: | Yang, Y, Arseni, D, Zhang, W, Huang, M, Lovestam, S.K.A, Schweighauser, M, Kotecha, A, Murzin, A.G, Peak-Chew, S.Y, Macdonald, J, Lavenir, I, Garringer, H.J, Gelpi, E, Newell, K.L, Kovacs, G.G, Vidal, R, Ghetti, B, Falcon, B, Scheres, S.H.W, Goedert, M. | | Deposit date: | 2021-11-01 | | Release date: | 2021-11-24 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Cryo-EM structures of amyloid-beta 42 filaments from human brains.
Science, 375, 2022
|
|
8YT1
 
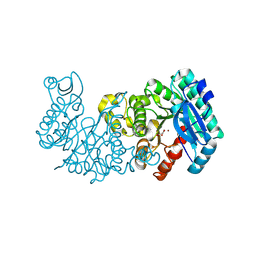 | |
8YT2
 
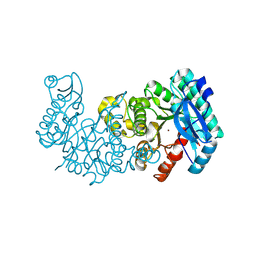 | | Crystal structure of ACMSD mutant W194A | | Descriptor: | 2-amino-3-carboxymuconate 6-semialdehyde decarboxylase, ZINC ION | | Authors: | Yang, Y, Liu, A. | | Deposit date: | 2024-03-24 | | Release date: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Crystal structure of ACMSD mutant W194A
To Be Published
|
|
5MN9
 
 | |
7Q4B
 
 | | Type I beta-amyloid 42 Filaments from Human Brain | | Descriptor: | Amyloid-beta precursor protein, UNKNOWN ATOM OR ION | | Authors: | Yang, Y, Arseni, D, Zhang, W, Huang, M, Lovestam, S.K.A, Schweighauser, M, Kotecha, A, Murzin, A.G, Peak-Chew, S.Y, Macdonald, J, Lavenir, I, Garringer, H.J, Gelpi, E, Newell, K.L, Kovacs, G.G, Vidal, R, Ghetti, B, Falcon, B, Scheres, S.H.W, Goedert, M. | | Deposit date: | 2021-10-30 | | Release date: | 2021-11-24 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Cryo-EM structures of amyloid-beta 42 filaments from human brains.
Science, 375, 2022
|
|
4S1Z
 
 | | Crystal structure of TRABID NZF1 in complex with K29 linked di-Ubiquitin | | Descriptor: | Ubiquitin, Ubiquitin thioesterase ZRANB1, ZINC ION | | Authors: | Kristariyanto, Y.A, Abdul Rehman, S.A, Campbell, D.G, Morrice, N.A, Johnson, C, Toth, R, Kulathu, Y. | | Deposit date: | 2015-01-16 | | Release date: | 2015-04-08 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.03 Å) | | Cite: | K29-selective ubiquitin binding domain reveals structural basis of specificity and heterotypic nature of k29 polyubiquitin.
Mol.Cell, 58, 2015
|
|
4S22
 
 | | Crystal structure of K29 linked di-Ubiquitin | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, IODIDE ION, ... | | Authors: | Kristariyanto, Y.A, Abdul Rehman, S.A, Campbell, D.G, Morrice, N.A, Johnson, C, Toth, R, Kulathu, Y. | | Deposit date: | 2015-01-17 | | Release date: | 2015-04-08 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | K29-selective ubiquitin binding domain reveals structural basis of specificity and heterotypic nature of k29 polyubiquitin.
Mol.Cell, 58, 2015
|
|
2RVJ
 
 | |
1UAW
 
 | | Solution structure of the N-terminal RNA-binding domain of mouse Musashi1 | | Descriptor: | mouse-musashi-1 | | Authors: | Miyanoiri, Y, Kobayashi, H, Watanabe, M, Ikeda, T, Nagata, T, Okano, H, Uesugi, S, Katahira, M. | | Deposit date: | 2003-03-24 | | Release date: | 2004-03-24 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Origin of higher affinity to RNA of the N-terminal RNA-binding domain than that of the C-terminal one of a mouse neural protein, musashi1, as revealed by comparison of their structures, modes of interaction, surface electrostatic potentials, and backbone dynamics
J.Biol.Chem., 278, 2003
|
|
4V0X
 
 | | The crystal structure of mouse PP1G in complex with truncated human PPP1R15B (631-684) | | Descriptor: | MANGANESE (II) ION, PROTEIN PHOSPHATASE 1 REGULATORY SUBUNIT 15B, PROTEIN PHOSPHATASE PP1-GAMMA CATALYTIC SUBUNIT | | Authors: | Chen, R, Yan, Y, Casado, A.C, Ron, D, Read, R.J. | | Deposit date: | 2014-09-18 | | Release date: | 2015-03-25 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | G-actin provides substrate-specificity to eukaryotic initiation factor 2 alpha holophosphatases.
Elife, 4, 2015
|
|
