5XBZ
 
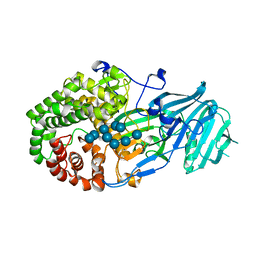 | | Crystal structure of GH family 81 beta-1,3-glucanase from Rhizomucr miehei complexed with laminaripentaose | | Descriptor: | Endo-beta-1,3-glucanase, beta-D-glucopyranose-(1-3)-beta-D-glucopyranose-(1-3)-beta-D-glucopyranose, beta-D-glucopyranose-(1-3)-beta-D-glucopyranose-(1-3)-beta-D-glucopyranose-(1-3)-beta-D-glucopyranose, ... | | Authors: | Yang, S, Qin, Z, Zhou, P, Yan, Q, Jiang, Z. | | Deposit date: | 2017-03-21 | | Release date: | 2018-03-28 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Catalytic mechanism of glycoside hydrolase family 81 beta-1,3-glucanase
To Be Published
|
|
5Z9S
 
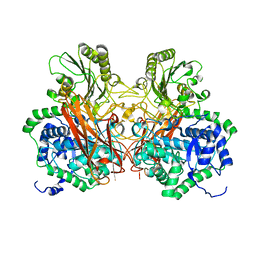 | |
2MPX
 
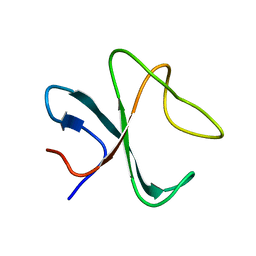 | |
2M02
 
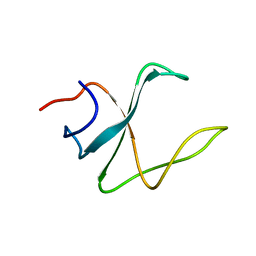 | | 3D structure of cap-gly domain of mammalian dynactin determined by magic angle spinning NMR spectroscopy | | Descriptor: | Dynactin subunit 1 | | Authors: | Yan, S, Hou, G, Schwieters, C.D, Ahmed, S, Williams, J.C, Polenova, T. | | Deposit date: | 2012-10-15 | | Release date: | 2013-05-08 | | Last modified: | 2024-05-15 | | Method: | SOLID-STATE NMR | | Cite: | Three-Dimensional Structure of CAP-Gly Domain of Mammalian Dynactin Determined by Magic Angle Spinning NMR Spectroscopy: Conformational Plasticity and Interactions with End-Binding Protein EB1.
J.Mol.Biol., 425, 2013
|
|
2IJN
 
 | | Isothiazoles as active-site inhibitors of HCV NS5B polymerase | | Descriptor: | (2R,3R)-3-{[3,5-BIS(TRIFLUOROMETHYL)PHENYL]AMINO}-2-CYANO-3-THIOXOPROPANAMIDE, RNA polymerase NS5B | | Authors: | Yan, S, Yao, N. | | Deposit date: | 2006-09-29 | | Release date: | 2006-11-28 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Isothiazoles as active-site inhibitors of HCV NS5B polymerase
Bioorg.Med.Chem.Lett., 17, 2007
|
|
3E8N
 
 | | X-ray structure of the human mitogen-activated protein kinase kinase 1 (MEK1) complexed with a potent inhibitor RDEA119 and MgATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Dual specificity mitogen-activated protein kinase kinase 1, MAGNESIUM ION, ... | | Authors: | Yan, S, Wang, Z.M. | | Deposit date: | 2008-08-20 | | Release date: | 2009-09-01 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | RDEA119/BAY 869766: a potent, selective, allosteric inhibitor of MEK1/2 for the treatment of cancer.
Cancer Res., 69, 2009
|
|
6LIG
 
 | |
4JL3
 
 | | Crystal structure of ms6564-dna complex | | Descriptor: | DNA (31-MER), Transcriptional regulator, TetR family | | Authors: | Yang, S.F, Gao, Z.Q, He, Z.G, Dong, Y.H. | | Deposit date: | 2013-03-12 | | Release date: | 2013-06-26 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis for interaction between Mycobacterium smegmatis Ms6564, a TetR family master regulator, and its target DNA.
J.Biol.Chem., 288, 2013
|
|
4JKZ
 
 | | Crystal structure of ms6564 from mycobacterium smegmatis | | Descriptor: | Transcriptional regulator, TetR family | | Authors: | Yang, S.F, Gao, Z.Q, He, Z.G, Dong, Y.H. | | Deposit date: | 2013-03-12 | | Release date: | 2013-06-26 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis for interaction between Mycobacterium smegmatis Ms6564, a TetR family master regulator, and its target DNA.
J.Biol.Chem., 288, 2013
|
|
6VNW
 
 | | Cryo-EM structure of apo-BBSome | | Descriptor: | BBS1 domain-containing protein, Bardet-Biedl syndrome 18 protein, Bardet-Biedl syndrome 2 protein homolog, ... | | Authors: | Yang, S, Walz, T, Nachury, M, Chou, H. | | Deposit date: | 2020-01-29 | | Release date: | 2020-07-01 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.44 Å) | | Cite: | Near-atomic structures of the BBSome reveal the basis for BBSome activation and binding to GPCR cargoes.
Elife, 9, 2020
|
|
6VOA
 
 | | Cryo-EM structure of the BBSome-ARL6 complex | | Descriptor: | ADP-ribosylation factor-like protein 6, BBS1 domain-containing protein, Bardet-Biedl syndrome 18 protein, ... | | Authors: | Yang, S, Walz, T, Nachury, M.V. | | Deposit date: | 2020-01-30 | | Release date: | 2020-06-24 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Near-atomic structures of the BBSome reveal the basis for BBSome activation and binding to GPCR cargoes.
Elife, 9, 2020
|
|
2KAP
 
 | | Solution structure of DLC1-SAM | | Descriptor: | Rho GTPase-activating protein 7 | | Authors: | Yang, S, Yang, D. | | Deposit date: | 2008-11-12 | | Release date: | 2009-10-20 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Characterization of DLC1-SAM equilibrium unfolding at the amino acid residue level
Biochemistry, 48, 2009
|
|
3L43
 
 | | Crystal structure of the dynamin 3 GTPase domain bound with GDP | | Descriptor: | Dynamin-3, GUANOSINE-5'-DIPHOSPHATE, UNKNOWN ATOM OR ION | | Authors: | Yang, S, Tempel, W, Tong, Y, Nedyalkova, L, Guan, X, Crombet, L, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Weigelt, J, Bochkarev, A, Park, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-12-18 | | Release date: | 2010-01-19 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Crystal structure of the dynamin 3 GTPase domain bound with GDP
to be published
|
|
2GYT
 
 | |
2KET
 
 | |
5JJY
 
 | | Crystal structure of SETD2 bound to histone H3.3 K36M peptide | | Descriptor: | Histone H3.3, Histone-lysine N-methyltransferase SETD2, S-ADENOSYL-L-HOMOCYSTEINE, ... | | Authors: | Yang, S, Zheng, X, Li, H. | | Deposit date: | 2016-04-25 | | Release date: | 2016-11-02 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.053 Å) | | Cite: | Molecular basis for oncohistone H3 recognition by SETD2 methyltransferase
Genes Dev., 30, 2016
|
|
3UI7
 
 | | Discovery of orally active pyrazoloquinoline as a potent PDE10 inhibitor for the management of schizophrenia | | Descriptor: | 6-methoxy-3,8-dimethyl-4-(morpholin-4-ylmethyl)-1H-pyrazolo[3,4-b]quinoline, MAGNESIUM ION, ZINC ION, ... | | Authors: | Yang, S, Smotryski, J, Mcelroy, W, Ho, G, Tulshian, D, Greenlee, W.J, Hodgson, R, Xiao, L, Hruza, A. | | Deposit date: | 2011-11-04 | | Release date: | 2011-12-21 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Discovery of orally active pyrazoloquinolines as potent PDE10 inhibitors for the management of schizophrenia.
Bioorg.Med.Chem.Lett., 22, 2012
|
|
6V0M
 
 | |
8XLC
 
 | |
8XKJ
 
 | |
8XLB
 
 | |
5XTY
 
 | |
8XKK
 
 | |
4GOY
 
 | | The crystal structure of human fascin 1 K41A mutant | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, BROMIDE ION, CHLORIDE ION, ... | | Authors: | Yang, S.Y, Huang, F.K, Huang, J, Chen, S, Jakoncic, J, Leo-Macias, A, Diaz-Avalos, R, Chen, L, Zhang, J.J, Huang, X.Y. | | Deposit date: | 2012-08-20 | | Release date: | 2012-11-28 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Molecular mechanism of fascin function in filopodial formation.
J.Biol.Chem., 288, 2013
|
|
4GOV
 
 | | The crystal structure of human fascin 1 S39D mutant | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, BROMIDE ION, CHLORIDE ION, ... | | Authors: | Yang, S.Y, Huang, F.K, Huang, J, Chen, S, Jakoncic, J, Leo-Macias, A, Diaz-Avalos, R, Chen, L, Zhang, J.J, Huang, X.Y. | | Deposit date: | 2012-08-20 | | Release date: | 2012-11-28 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Molecular mechanism of fascin function in filopodial formation.
J.Biol.Chem., 288, 2013
|
|
