6W5T
 
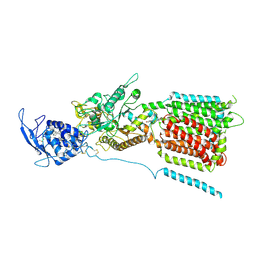 | | NPC1 structure in GDN micelles at pH 5.5, conformation a | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHOLESTEROL, ... | | Authors: | Yan, N, Qian, H.W, Wu, X.L. | | Deposit date: | 2020-03-13 | | Release date: | 2020-06-17 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structural Basis of Low-pH-Dependent Lysosomal Cholesterol Egress by NPC1 and NPC2.
Cell, 182, 2020
|
|
6W5U
 
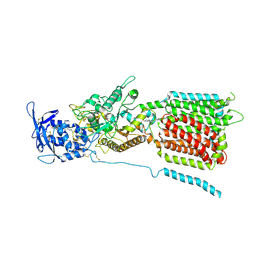 | | NPC1 structure in GDN micelles at pH 5.5, conformation b | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHOLESTEROL, ... | | Authors: | Yan, N, Qian, H.W, Wu, X.L. | | Deposit date: | 2020-03-13 | | Release date: | 2020-06-17 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structural Basis of Low-pH-Dependent Lysosomal Cholesterol Egress by NPC1 and NPC2.
Cell, 182, 2020
|
|
6W5V
 
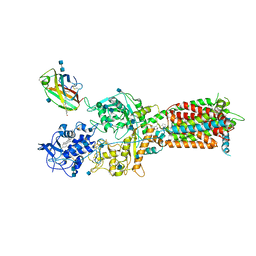 | | NPC1-NPC2 complex structure at pH 5.5 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHOLESTEROL, ... | | Authors: | Yan, N, Qian, H.W, Wu, X.L. | | Deposit date: | 2020-03-13 | | Release date: | 2020-06-17 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structural Basis of Low-pH-Dependent Lysosomal Cholesterol Egress by NPC1 and NPC2.
Cell, 182, 2020
|
|
6VX3
 
 | | NaChBac in GDN | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, BH1501 protein | | Authors: | Yan, N, Gao, S. | | Deposit date: | 2020-02-21 | | Release date: | 2020-06-24 | | Last modified: | 2020-07-08 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Employing NaChBac for cryo-EM analysis of toxin action on voltage-gated Na+channels in nanodisc.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6W6O
 
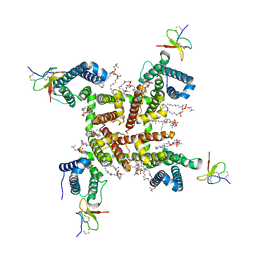 | | NaChBac-Nav1.7VSDII chimera and HWTX-IV complex | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, Huwentoxin-IV, NaChBac-Nav1.7VSDII chimera | | Authors: | Yan, N, Gao, S. | | Deposit date: | 2020-03-17 | | Release date: | 2020-06-24 | | Last modified: | 2021-01-13 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Employing NaChBac for cryo-EM analysis of toxin action on voltage-gated Na + channels in nanodisc.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6VWX
 
 | | NaChBac in lipid nanodisc | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, BH1501 protein, SODIUM ION | | Authors: | Yan, N, Gao, S. | | Deposit date: | 2020-02-20 | | Release date: | 2020-06-24 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Employing NaChBac for cryo-EM analysis of toxin action on voltage-gated Na+channels in nanodisc.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6VXO
 
 | | NaChBac-Nav1.7VSDII chimera in nanodisc | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, NaChBac-Nav1.7VSDII chimera | | Authors: | Yan, N, Gao, S. | | Deposit date: | 2020-02-22 | | Release date: | 2020-06-24 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Employing NaChBac for cryo-EM analysis of toxin action on voltage-gated Na+channels in nanodisc.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6W4Y
 
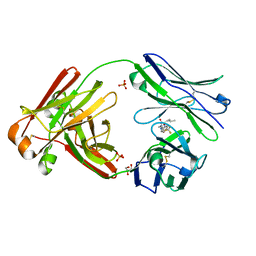 | | Structure of full-length human lambda-6A light chain JTO in complex with hydantoin stabilizer | | Descriptor: | 2-[(4~{R})-4-(2-methylpropyl)-2,5-bis(oxidanylidene)imidazolidin-1-yl]-~{N}-[4-(trifluoromethyl)phenyl]ethanamide, GLYCEROL, JTO light chain, ... | | Authors: | Yan, N.L, Morgan, G.J, Kelly, J.W. | | Deposit date: | 2020-03-11 | | Release date: | 2020-07-01 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Structural basis for the stabilization of amyloidogenic immunoglobulin light chains by hydantoins.
Bioorg.Med.Chem.Lett., 30, 2020
|
|
7MIX
 
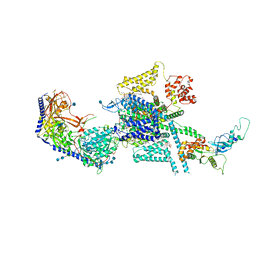 | | Human N-type voltage-gated calcium channel Cav2.2 in the presence of ziconotide at 3.0 Angstrom resolution | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Yan, N, Gao, S, Yao, X. | | Deposit date: | 2021-04-18 | | Release date: | 2021-07-07 | | Last modified: | 2021-08-18 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structure of human Ca v 2.2 channel blocked by the painkiller ziconotide.
Nature, 596, 2021
|
|
7MIY
 
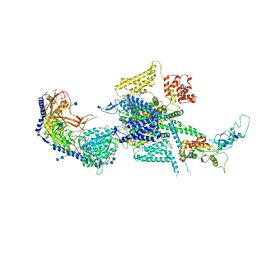 | | Human N-type voltage-gated calcium channel Cav2.2 at 3.1 Angstrom resolution | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Yan, N, Gao, S, Yao, X. | | Deposit date: | 2021-04-18 | | Release date: | 2021-07-07 | | Last modified: | 2021-08-18 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structure of human Ca v 2.2 channel blocked by the painkiller ziconotide.
Nature, 596, 2021
|
|
7JPX
 
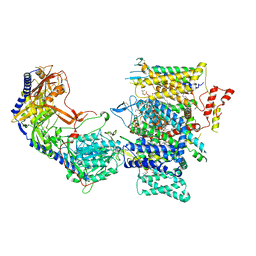 | | Rabbit Cav1.1 in the presence of 100 micromolar amlodipine in nanodiscs at 2.9 Angstrom resolution | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOETHANOLAMINE, ... | | Authors: | Yan, N, Gao, S. | | Deposit date: | 2020-08-10 | | Release date: | 2020-11-18 | | Last modified: | 2021-03-10 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural Basis of the Modulation of the Voltage-Gated Calcium Ion Channel Ca v 1.1 by Dihydropyridine Compounds*.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
7LMN
 
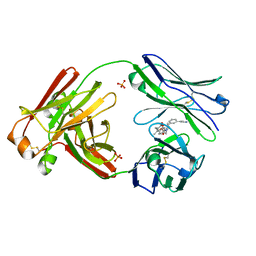 | |
7JPV
 
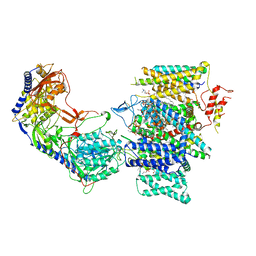 | | Rabbit Cav1.1 in the presence of 1 micromolar (S)-(-)-Bay K8644 in nanodiscs at 3.4 Angstrom resolution | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOETHANOLAMINE, CALCIUM ION, ... | | Authors: | Yan, N, Gao, S. | | Deposit date: | 2020-08-10 | | Release date: | 2020-11-18 | | Last modified: | 2021-03-10 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural Basis of the Modulation of the Voltage-Gated Calcium Ion Channel Ca v 1.1 by Dihydropyridine Compounds*.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
6DMB
 
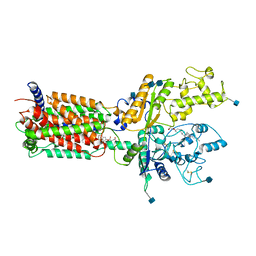 | | Cryo-EM structure of human Ptch1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHOLESTEROL HEMISUCCINATE, ... | | Authors: | Yan, N, Gong, X, Qian, H.W. | | Deposit date: | 2018-06-04 | | Release date: | 2018-07-11 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structural basis for the recognition of Sonic Hedgehog by human Patched1.
Science, 361, 2018
|
|
7LMO
 
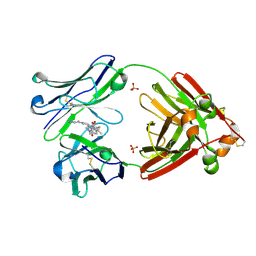 | | Structure of full-length human lambda-6A light chain JTO in complex with stabilizer 34 [3-(2-(7-(diethylamino)-4-methyl-2-oxo-2H-chromen-3-yl)ethyl)-7-(1H-imidazole-5-carbonyl)-1,3,7-triazaspiro[4.4]nonane-2,4-dione] | | Descriptor: | (5~{R})-3-[2-[7-(diethylamino)-4-methyl-2-oxidanylidene-chromen-3-yl]ethyl]-7-(1~{H}-imidazol-4-ylcarbonyl)-1,3,7-triazaspiro[4.4]nonane-2,4-dione, JTO light chain, PHOSPHATE ION | | Authors: | Yan, N.L, Wilson, I.A, Kelly, J.W. | | Deposit date: | 2021-02-05 | | Release date: | 2021-05-19 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Discovery of Potent Coumarin-Based Kinetic Stabilizers of Amyloidogenic Immunoglobulin Light Chains Using Structure-Based Design.
J.Med.Chem., 64, 2021
|
|
7LMR
 
 | |
7LMP
 
 | | Structure of full-length human lambda-6A light chain JTO in complex with stabilizer 36 [3-(2-(7-(diethylamino)-4-methyl-2-oxo-2H-chromen-3-yl)ethyl)-8-(1H-imidazole-4-carbonyl)-1,3,8-triazaspiro[4.5]decane-2,4-dione] | | Descriptor: | 3-[2-[7-(diethylamino)-4-methyl-2-oxidanylidene-chromen-3-yl]ethyl]-8-(1~{H}-imidazol-5-ylcarbonyl)-1,3,8-triazaspiro[4.5]decane-2,4-dione, JTO light chain, PHOSPHATE ION | | Authors: | Yan, N.L, Wilson, I.A, Kelly, J.W. | | Deposit date: | 2021-02-05 | | Release date: | 2021-05-19 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Discovery of Potent Coumarin-Based Kinetic Stabilizers of Amyloidogenic Immunoglobulin Light Chains Using Structure-Based Design.
J.Med.Chem., 64, 2021
|
|
7LMQ
 
 | |
1SE0
 
 | | Crystal structure of DIAP1 BIR1 bound to a Grim peptide | | Descriptor: | Apoptosis 1 inhibitor, Cell death protein Grim, ZINC ION | | Authors: | Yan, N, Wu, J.W, Shi, Y. | | Deposit date: | 2004-02-15 | | Release date: | 2004-04-27 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Molecular mechanisms of DrICE inhibition by DIAP1 and removal of inhibition by Reaper, Hid and Grim.
Nat.Struct.Mol.Biol., 11, 2004
|
|
1TY4
 
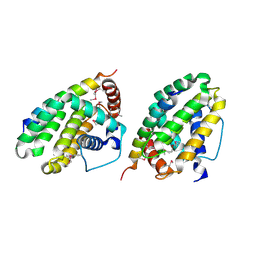 | | Crystal structure of a CED-9/EGL-1 complex | | Descriptor: | Apoptosis regulator ced-9, EGg Laying defective EGL-1, programmed cell death activator | | Authors: | Yan, N, Gu, L, Kokel, D, Xue, D, Shi, Y. | | Deposit date: | 2004-07-07 | | Release date: | 2004-09-28 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural, Biochemical, and Functional Analyses of CED-9 Recognition by the Proapoptotic Proteins EGL-1 and CED-4
Mol.Cell, 15, 2004
|
|
1SDZ
 
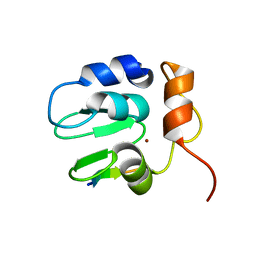 | | Crystal structure of DIAP1 BIR1 bound to a Reaper peptide | | Descriptor: | Apoptosis 1 inhibitor, Reaper, ZINC ION | | Authors: | Yan, N, Wu, J.W, Shi, Y. | | Deposit date: | 2004-02-15 | | Release date: | 2004-04-27 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Molecular mechanisms of DrICE inhibition by DIAP1 and removal of inhibition by Reaper, Hid and Grim.
Nat.Struct.Mol.Biol., 11, 2004
|
|
8FO3
 
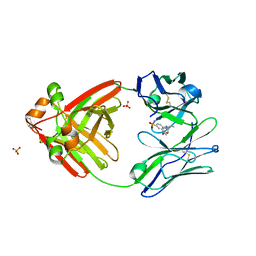 | |
8FO4
 
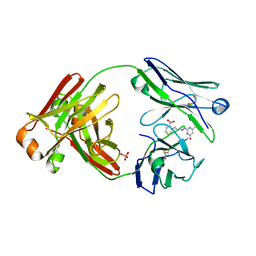 | |
8FO5
 
 | |
2FP3
 
 | |
