7OK0
 
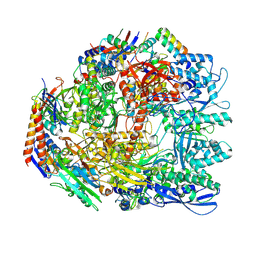 | | Cryo-EM structure of the Sulfolobus acidocaldarius RNA polymerase at 2.88 A | | Descriptor: | Conserved protein, DNA-directed RNA polymerase subunit A', DNA-directed RNA polymerase subunit A'', ... | | Authors: | Pilotto, S, Fouqueau, T, Lukoyanova, N, Sheppard, C, Lucas-Staat, S, Diaz-Santin, L.M, Matelska, D, Prangishvili, D, Cheung, A.C.M, Werner, F. | | Deposit date: | 2021-05-17 | | Release date: | 2021-08-25 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural basis of RNA polymerase inhibition by viral and host factors.
Nat Commun, 12, 2021
|
|
7OQY
 
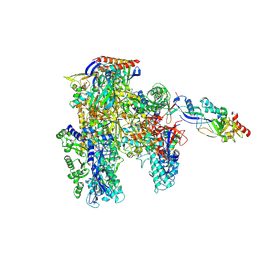 | | Cryo-EM structure of the cellular negative regulator TFS4 bound to the archaeal RNA polymerase | | Descriptor: | Conserved protein, DNA-directed RNA polymerase subunit A', DNA-directed RNA polymerase subunit A'', ... | | Authors: | Pilotto, S, Fouqueau, T, Lukoyanova, N, Sheppard, C, Lucas-Staat, S, Diaz-Santin, L.M, Matelska, D, Prangishvili, D, Cheung, A.C.M, Werner, F. | | Deposit date: | 2021-06-04 | | Release date: | 2021-08-25 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (2.61 Å) | | Cite: | Structural basis of RNA polymerase inhibition by viral and host factors.
Nat Commun, 12, 2021
|
|
7OQ4
 
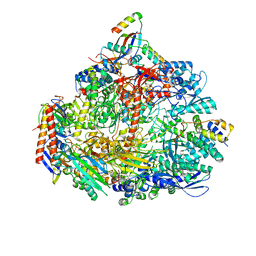 | | Cryo-EM structure of the ATV RNAP Inhibitory Protein (RIP) bound to the DNA-binding channel of the host's RNA polymerase | | Descriptor: | Conserved protein, DNA-directed RNA polymerase subunit A', DNA-directed RNA polymerase subunit A'', ... | | Authors: | Pilotto, S, Fouqueau, T, Lukoyanova, N, Sheppard, C, Lucas-Staat, S, Diaz-Santin, L.M, Matelska, D, Prangishvili, D, Cheung, A.C.M, Werner, F. | | Deposit date: | 2021-06-02 | | Release date: | 2021-08-25 | | Last modified: | 2021-10-13 | | Method: | ELECTRON MICROSCOPY (3.27 Å) | | Cite: | Structural basis of RNA polymerase inhibition by viral and host factors.
Nat Commun, 12, 2021
|
|
4JJ4
 
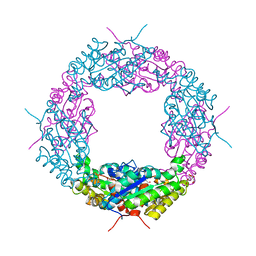 | | Crystal structure of a catalytic mutant of Axe2 (Axe2-D191A), an acetylxylan esterase from Geobacillus stearothermophilus | | Descriptor: | Acetyl xylan esterase, CHLORIDE ION, GLYCEROL | | Authors: | Lansky, S, Alalouf, O, Solomon, V, Alhassid, A, Belrahli, H, Govada, L, Chayan, N.E, Shoham, Y, Shoham, G. | | Deposit date: | 2013-03-07 | | Release date: | 2014-03-12 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | To be published
To be Published
|
|
1T72
 
 | | Crystal structure of phosphate transport system protein phoU from Aquifex aeolicus | | Descriptor: | Phosphate transport system protein phoU homolog | | Authors: | Oganesyan, V, Kim, S.-H, Oganesyan, N, Jancarik, J, Adams, P.D, Kim, R, Berkeley Structural Genomics Center (BSGC) | | Deposit date: | 2004-05-07 | | Release date: | 2004-12-07 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structure of the "PhoU-like" phosphate uptake regulator from Aquifex aeolicus.
J.Bacteriol., 187, 2005
|
|
1MFJ
 
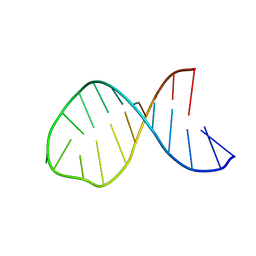 | |
6JQN
 
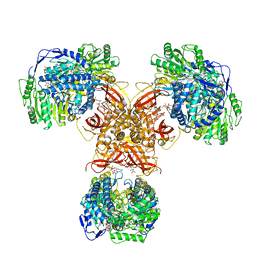 | | Structure of PaaZ, a bifunctional enzyme in complex with NADP+ and OCoA | | Descriptor: | Bifunctional protein PaaZ, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, OCTANOYL-COENZYME A | | Authors: | Gakher, L, Vinothkumar, K.R, Katagihallimath, N, Sowdhamini, R, Sathyanarayanan, N, Cannone, G. | | Deposit date: | 2019-03-31 | | Release date: | 2019-09-11 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Molecular basis for metabolite channeling in a ring opening enzyme of the phenylacetate degradation pathway.
Nat Commun, 10, 2019
|
|
6JQO
 
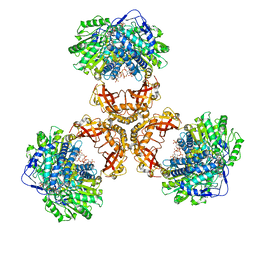 | | Structure of PaaZ, a bifunctional enzyme in complex with NADP+ and CCoA | | Descriptor: | Bifunctional protein PaaZ, CROTONYL COENZYME A, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Gakher, L, Vinothkumar, K.R, Katagihallimath, N, Sowdhamini, R, Sathyanarayanan, N, Cannone, G. | | Deposit date: | 2019-03-31 | | Release date: | 2019-09-11 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Molecular basis for metabolite channeling in a ring opening enzyme of the phenylacetate degradation pathway.
Nat Commun, 10, 2019
|
|
6JQL
 
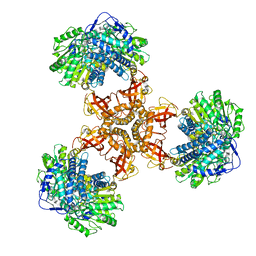 | | Structure of PaaZ, a bifunctional enzyme | | Descriptor: | Bifunctional protein PaaZ | | Authors: | Gakher, L, Vinothkumar, K.R, Katagihallimath, N, Sowdhamini, R, Sathyanarayanan, N, Cannone, G. | | Deposit date: | 2019-03-31 | | Release date: | 2019-09-11 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Molecular basis for metabolite channeling in a ring opening enzyme of the phenylacetate degradation pathway.
Nat Commun, 10, 2019
|
|
6JQM
 
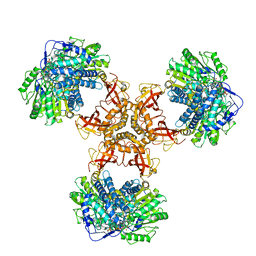 | | Structure of PaaZ with NADPH | | Descriptor: | Bifunctional protein PaaZ, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Gakher, L, Vinothkumar, K.R, Katagihallimath, N, Sowdhamini, R, Sathyanarayanan, N, Cannone, G. | | Deposit date: | 2019-03-31 | | Release date: | 2019-09-11 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Molecular basis for metabolite channeling in a ring opening enzyme of the phenylacetate degradation pathway.
Nat Commun, 10, 2019
|
|
6I9D
 
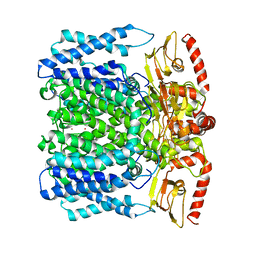 | | MloK1 consensus structure from single particle analysis of 2D crystals | | Descriptor: | Cyclic nucleotide-gated potassium channel mll3241, POTASSIUM ION | | Authors: | Righetto, R, Biyani, N, Kowal, J, Chami, M, Stahlberg, H. | | Deposit date: | 2018-11-23 | | Release date: | 2019-04-24 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Retrieving high-resolution information from disordered 2D crystals by single-particle cryo-EM.
Nat Commun, 10, 2019
|
|
6IAX
 
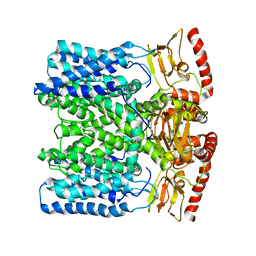 | | MloK1 model from single particle analysis of 2D crystals, class 1 (extended conformation) | | Descriptor: | Cyclic nucleotide-gated potassium channel mll3241, POTASSIUM ION | | Authors: | Righetto, R, Biyani, N, Kowal, J, Chami, M, Stahlberg, H. | | Deposit date: | 2018-11-27 | | Release date: | 2019-04-24 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (5.2 Å) | | Cite: | Retrieving high-resolution information from disordered 2D crystals by single-particle cryo-EM.
Nat Commun, 10, 2019
|
|
7NST
 
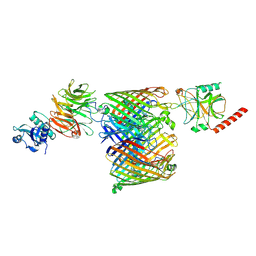 | | ColicinE9 partial translocation complex | | Descriptor: | Colicin-E9, Outer membrane protein F, Tol-Pal system protein TolB | | Authors: | Webby, M.N, Kleanthous, C, Lukoyanova, N, Housden, N.G. | | Deposit date: | 2021-03-08 | | Release date: | 2021-08-11 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Porin threading drives receptor disengagement and establishes active colicin transport through Escherichia coli OmpF.
Embo J., 40, 2021
|
|
7NSU
 
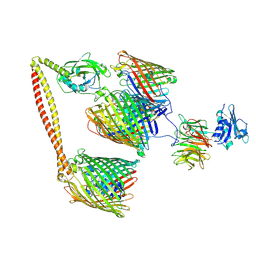 | | ColicinE9 intact translocation complex | | Descriptor: | Colicin-E9, Outer membrane protein F, Tol-Pal system protein TolB, ... | | Authors: | Webby, M.N, Kleanthous, C, Lukoyanova, N, Housden, N.G. | | Deposit date: | 2021-03-08 | | Release date: | 2021-08-11 | | Last modified: | 2021-11-17 | | Method: | ELECTRON MICROSCOPY (4.7 Å) | | Cite: | Porin threading drives receptor disengagement and establishes active colicin transport through Escherichia coli OmpF.
Embo J., 40, 2021
|
|
5FMW
 
 | | The poly-C9 component of the Complement Membrane Attack Complex | | Descriptor: | POLYC9 | | Authors: | Dudkina, N.V, Spicer, B.A, Reboul, C.F, Conroy, P.J, Lukoyanova, N, Elmlund, H, Law, R.H.P, Ekkel, S.M, Kondos, S.C, Goode, R.J.A, Ramm, G, Whisstock, J.C, Saibil, H.R, Dunstone, M.A. | | Deposit date: | 2015-11-10 | | Release date: | 2016-02-17 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (6.7 Å) | | Cite: | Structure of the Poly-C9 Component of the Complement Membrane Attack Complex
Nat.Commun., 7, 2016
|
|
6KK8
 
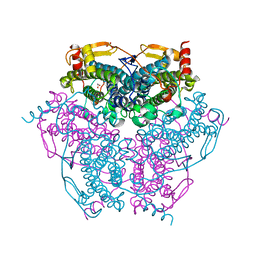 | | XN joint refinement of manganese catalase from Thermus Thermophilus HB27 | | Descriptor: | 1,2-ETHANEDIOL, MANGANESE (III) ION, OXYGEN ATOM, ... | | Authors: | Yamada, T, Yano, N, Kusaka, K. | | Deposit date: | 2019-07-24 | | Release date: | 2019-09-04 | | Last modified: | 2024-04-03 | | Method: | NEUTRON DIFFRACTION (1.37 Å), X-RAY DIFFRACTION | | Cite: | Single-crystal time-of-flight neutron Laue methods: application to manganese catalase from Thermus thermophilus HB27
J.Appl.Crystallogr., 2019
|
|
2M8K
 
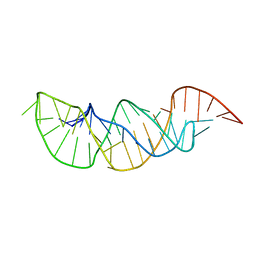 | | A pyrimidine motif triple helix in the Kluyveromyces lactis telomerase RNA pseudoknot is essential for function in vivo | | Descriptor: | RNA (48-MER) | | Authors: | Cash, D.D, Cohen, O, Kim, N, Shefer, K, Brown, Y, Ulyanov, N.B, Tzfati, Y, Feigon, J. | | Deposit date: | 2013-05-22 | | Release date: | 2013-06-19 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Pyrimidine motif triple helix in the Kluyveromyces lactis telomerase RNA pseudoknot is essential for function in vivo.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
6CBU
 
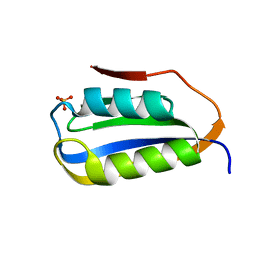 | | Crystal structure of C4S3: A computationally designed immunogen to target Carbohydrate-Occluded Epitopes on the HIV envelope | | Descriptor: | Acylphosphatase-1, SULFATE ION | | Authors: | Zhu, C, Ke, H.M, Swanstrom, R, Dokholyan, N.V. | | Deposit date: | 2018-02-05 | | Release date: | 2019-02-06 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Rationally designed carbohydrate-occluded epitopes elicit HIV-1 Env-specific antibodies.
Nat Commun, 10, 2019
|
|
6CFE
 
 | |
1R7W
 
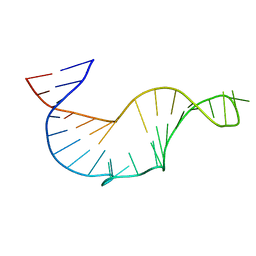 | | NMR STRUCTURE OF THE R(GGAGGACAUCCCUCACGGGUGACCGUGGUCCUCC), DOMAIN IV STEM-LOOP B OF ENTEROVIRAL IRES WITH AUCCCU BULGE | | Descriptor: | 34-MER | | Authors: | Du, Z, Ulyanov, N.B, Yu, J, James, T.L. | | Deposit date: | 2003-10-22 | | Release date: | 2004-05-25 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | NMR Structures of Loop B RNAs from the Stem-Loop IV Domain of the Enterovirus Internal Ribosome Entry Site: A Single C to U Substitution Drastically Changes the Shape and Flexibility of RNA(,).
Biochemistry, 43, 2004
|
|
1R7Z
 
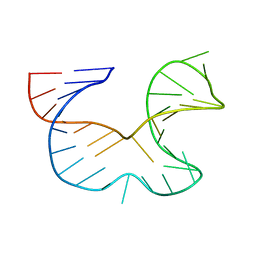 | | NMR STRUCTURE OF THE R(GGAGGACAUUCCUCACGGGUGACCGUGGUCCUCC), DOMAIN IV STEM-LOOP B OF ENTEROVIRAL IRES WITH AUUCCU BULGE | | Descriptor: | 34-MER | | Authors: | Du, Z, Ulyanov, N.B, Yu, J, James, T.L. | | Deposit date: | 2003-10-22 | | Release date: | 2004-05-25 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | NMR Structures of Loop B RNAs from the Stem-Loop IV Domain of the Enterovirus Internal Ribosome Entry Site: A Single C to U Substitution Drastically Changes the Shape and Flexibility of RNA(,).
Biochemistry, 43, 2004
|
|
1Z14
 
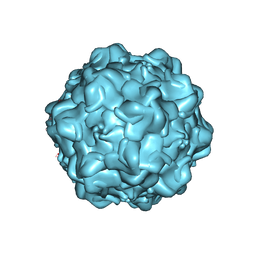 | | Structural Determinants of Tissue Tropism and In Vivo Pathogenicity for the Parvovirus Minute Virus of Mice | | Descriptor: | VP2 | | Authors: | Kontou, M, Govindasamy, L, Nam, H.J, Bryant, N, Llamas-Saiz, A.L, Foces-Foces, C, Hernando, E, Rubio, M.P, McKenna, R, Almendral, J.M, Agbandje-McKenna, M. | | Deposit date: | 2005-03-03 | | Release date: | 2005-09-06 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Structural determinants of tissue tropism and in vivo pathogenicity for the parvovirus minute virus of mice.
J.Virol., 79, 2005
|
|
1Z1C
 
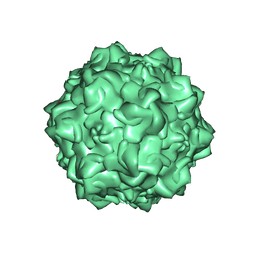 | | Structural Determinants of Tissue Tropism and In Vivo Pathogenicity for the Parvovirus Minute virus of Mice | | Descriptor: | 2'-DEOXYADENOSINE-5'-MONOPHOSPHATE, 5'-D(*AP*CP*AP*CP*CP*AP*AP*AP*A)-3', 5'-D(*AP*TP*CP*CP*TP*CP*TP*AP*TP*CP*AP*C)-3', ... | | Authors: | Kontou, M, Govindasamy, L, Nam, H.J, Bryant, N, Llamas-Saiz, A.L, Foces-Foces, C, Hernando, E, Rubio, M.P, McKenna, R, Almendral, J.M, Agbandje-McKenna, M. | | Deposit date: | 2005-03-03 | | Release date: | 2005-09-06 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structural determinants of tissue tropism and in vivo pathogenicity for the parvovirus minute virus of mice.
J.Virol., 79, 2005
|
|
6CFL
 
 | | Crystal structure of the Thermus thermophilus 70S ribosome in complex with lysyl-CAM and bound to protein Y (YfiA) at 2.6A resolution | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 16S Ribosomal RNA, 23S Ribosomal RNA, ... | | Authors: | Tereshchenkov, A.G, Dobosz-Bartoszek, M, Osterman, I.A, Marks, J, Sergeeva, V.A, Kasatsky, P, Komarova, E.S, Stavrianidi, A.N, Rodin, I.A, Konevega, A.L, Sergiev, P.V, Sumbatyan, N.V, Mankin, A.S, Bogdanov, A.A, Polikanov, Y.S. | | Deposit date: | 2018-02-15 | | Release date: | 2018-03-07 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Binding and Action of Amino Acid Analogs of Chloramphenicol upon the Bacterial Ribosome.
J. Mol. Biol., 430, 2018
|
|
6CFJ
 
 | | Crystal structure of the Thermus thermophilus 70S ribosome in complex with histidyl-CAM and bound to mRNA and A-, P-, and E-site tRNAs at 2.8A resolution | | Descriptor: | 16S Ribosomal RNA, 23S Ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Tereshchenkov, A.G, Dobosz-Bartoszek, M, Osterman, I.A, Marks, J, Sergeeva, V.A, Kasatsky, P, Komarova, E.S, Stavrianidi, A.N, Rodin, I.A, Konevega, A.L, Sergiev, P.V, Sumbatyan, N.V, Mankin, A.S, Bogdanov, A.A, Polikanov, Y.S. | | Deposit date: | 2018-02-15 | | Release date: | 2018-03-07 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Binding and Action of Amino Acid Analogs of Chloramphenicol upon the Bacterial Ribosome.
J. Mol. Biol., 430, 2018
|
|
