4GUR
 
 | | Crystal structure of LSD2-NPAC with H3 in space group P21 | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, Histone H3.3, ... | | Authors: | Chen, F, Dong, Z, Fang, J, Yang, Y, Li, Z, Xu, Y, Yang, H, Wang, P, Fang, R, Shi, Y, Xu, Y. | | Deposit date: | 2012-08-29 | | Release date: | 2013-01-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.506 Å) | | Cite: | LSD2/KDM1B and its cofactor NPAC/GLYR1 endow a structural and molecular model for regulation of H3K4 demethylation
Mol.Cell, 49, 2013
|
|
3MQ7
 
 | | Crystal Structure of Ectodomain Mutant of BST-2/Tetherin/CD317 | | Descriptor: | Bone marrow stromal antigen 2, CALCIUM ION | | Authors: | Xiong, Y, Yang, H, Wang, J, Meng, W. | | Deposit date: | 2010-04-27 | | Release date: | 2010-10-27 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Structural insight into the mechanisms of enveloped virus tethering by tetherin.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
2RJE
 
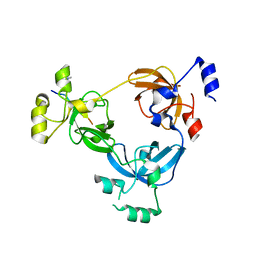 | | Crystal structure of L3MBTL1 in complex with H4K20Me2 (residues 17-25), orthorhombic form II | | Descriptor: | CHLORIDE ION, Histone H4, Lethal(3)malignant brain tumor-like protein | | Authors: | Allali-Hassani, A, Liu, Y, Herzanych, N, Ouyang, H, Mackenzie, F, Crombet, L, Loppnau, P, Kozieradzki, I, Vedadi, M, Weigelt, J, Sundstrom, M, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Min, J.R, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-10-14 | | Release date: | 2007-10-30 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | L3MBTL1 recognition of mono- and dimethylated histones.
Nat.Struct.Mol.Biol., 14, 2007
|
|
8E40
 
 | | Full-length APOBEC3G in complex with HIV-1 Vif, CBF-beta, and fork RNA | | Descriptor: | Core-binding factor subunit beta, DNA dC->dU-editing enzyme APOBEC-3G, RNA, ... | | Authors: | Ito, F, Alvarez-Cabrera, A.L, Liu, S, Yang, H, Shiriaeva, A, Zhou, Z.H, Chen, X.S. | | Deposit date: | 2022-08-17 | | Release date: | 2023-01-11 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.57 Å) | | Cite: | Structural basis for HIV-1 antagonism of host APOBEC3G via Cullin E3 ligase.
Sci Adv, 9, 2023
|
|
5K81
 
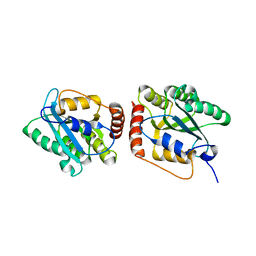 | | Crystal Structure of a Primate APOBEC3G N-Terminal Domain | | Descriptor: | Apolipoprotein B mRNA editing enzyme, catalytic peptide-like 3G,Apolipoprotein B mRNA editing enzyme, catalytic peptide-like 3G, ... | | Authors: | Xiao, X, Li, S.-X, Yang, H, Chen, X.S. | | Deposit date: | 2016-05-27 | | Release date: | 2016-08-10 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.006 Å) | | Cite: | Crystal structures of APOBEC3G N-domain alone and its complex with DNA.
Nat Commun, 7, 2016
|
|
5K82
 
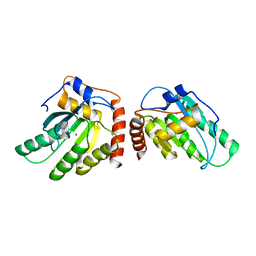 | | Crystal Structure of a Primate APOBEC3G N-Terminal Domain | | Descriptor: | Apolipoprotein B mRNA editing enzyme, catalytic peptide-like 3G,Apolipoprotein B mRNA editing enzyme, catalytic peptide-like 3G, ... | | Authors: | Xiao, X, Li, S.-X, Yang, H, Chen, X.S. | | Deposit date: | 2016-05-27 | | Release date: | 2016-08-10 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.913 Å) | | Cite: | Crystal structures of APOBEC3G N-domain alone and its complex with DNA.
Nat Commun, 7, 2016
|
|
1LBV
 
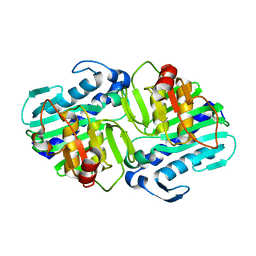 | | Crystal Structure of apo-form (P21) of dual activity FBPase/IMPase (AF2372) from Archaeoglobus fulgidus | | Descriptor: | fructose 1,6-bisphosphatase/inositol monophosphatase | | Authors: | Stieglitz, K.A, Johnson, K.A, Yang, H, Roberts, M.F, Seaton, B.A, Head, J.F, Stec, B. | | Deposit date: | 2002-04-04 | | Release date: | 2002-05-22 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of a dual activity IMPase/FBPase (AF2372) from Archaeoglobus fulgidus. The story of a mobile loop.
J.Biol.Chem., 277, 2002
|
|
7A5F
 
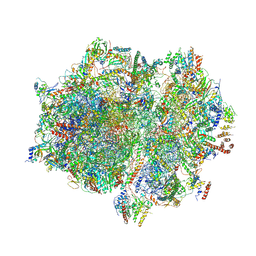 | | Structure of the stalled human mitoribosome with P- and E-site mt-tRNAs | | Descriptor: | 12S rRNA, 16S rRNA, 28S ribosomal protein S10, ... | | Authors: | Desai, N, Yang, H, Chandrasekaran, V, Kazi, R, Minczuk, M, Ramakrishnan, V. | | Deposit date: | 2020-08-21 | | Release date: | 2020-12-23 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Elongational stalling activates mitoribosome-associated quality control.
Science, 370, 2020
|
|
7A5H
 
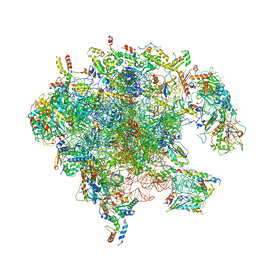 | | Structure of the split human mitoribosomal large subunit with rescue factors mtRF-R and MTRES1 | | Descriptor: | 16S rRNA, 39S ribosomal protein L10, mitochondrial, ... | | Authors: | Desai, N, Yang, H, Chandrasekaran, V, Kazi, R, Minczuk, M, Ramakrishnan, V. | | Deposit date: | 2020-08-21 | | Release date: | 2020-12-23 | | Last modified: | 2023-03-01 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Elongational stalling activates mitoribosome-associated quality control.
Science, 370, 2020
|
|
7A5I
 
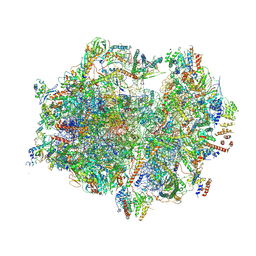 | | Structure of the human mitoribosome with A- P-and E-site mt-tRNAs | | Descriptor: | 12S rRNA, 16S rRNA, 28S ribosomal protein S10, ... | | Authors: | Desai, N, Yang, H, Chandrasekaran, V, Kazi, R, Minczuk, M, Ramakrishnan, V. | | Deposit date: | 2020-08-21 | | Release date: | 2020-12-23 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Elongational stalling activates mitoribosome-associated quality control.
Science, 370, 2020
|
|
1LBW
 
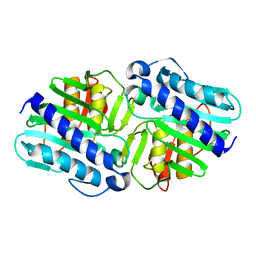 | | Crystal Structure of apo-form (P32) of dual activity FBPase/IMPase (AF2372) from Archaeoglobus fulgidus | | Descriptor: | fructose 1,6-bisphosphatase/inositol monophosphatase | | Authors: | Stieglitz, K.A, Johnson, K.A, Yang, H, Roberts, M.F, Seaton, B.A, Head, J.F, Stec, B. | | Deposit date: | 2002-04-04 | | Release date: | 2002-05-22 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of a dual activity IMPase/FBPase (AF2372) from Archaeoglobus fulgidus. The story of a mobile loop.
J.Biol.Chem., 277, 2002
|
|
1HMU
 
 | | ACTIVE SITE OF CHONDROITINASE AC LYASE REVEALED BY THE STRUCTURE OF ENZYME-OLIGOSACCHARIDE COMPLEXES AND MUTAGENESIS | | Descriptor: | 2-O-methyl-beta-L-fucopyranose-(1-4)-beta-D-xylopyranose-(1-4)-alpha-D-glucopyranuronic acid-(1-2)-[alpha-L-rhamnopyranose-(1-4)]alpha-D-mannopyranose, 4-deoxy-alpha-L-threo-hex-4-enopyranuronic acid-(1-3)-2-acetamido-2-deoxy-4-O-sulfo-beta-D-galactopyranose, CALCIUM ION, ... | | Authors: | Huang, W, Boju, L, Tkalec, L, Su, H, Yang, H.O, Gunay, N.S, Linhardt, R.J, Kim, Y.S, Matte, A, Cygler, M. | | Deposit date: | 2000-12-05 | | Release date: | 2001-05-02 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Active site of chondroitin AC lyase revealed by the structure of enzyme-oligosaccharide complexes and mutagenesis.
Biochemistry, 40, 2001
|
|
1HMW
 
 | | ACTIVE SITE OF CHONDROITINASE AC LYASE REVEALED BY THE STRUCTURE OF ENZYME-OLIGOSACCHARIDE COMPLEXES AND MUTAGENESIS | | Descriptor: | 2-O-methyl-beta-L-fucopyranose-(1-4)-beta-D-xylopyranose-(1-4)-alpha-D-glucopyranuronic acid-(1-2)-[alpha-L-rhamnopyranose-(1-4)]alpha-D-mannopyranose, 4-deoxy-alpha-L-threo-hex-4-enopyranuronic acid-(1-3)-2-acetamido-2-deoxy-4-O-sulfo-beta-D-galactopyranose-(1-4)-beta-D-glucopyranuronic acid-(1-3)-2-acetamido-2-deoxy-6-O-sulfo-beta-D-galactopyranose, CALCIUM ION, ... | | Authors: | Huang, W, Boju, L, Tkalec, L, Su, H, Yang, H.O, Gunay, N.S, Linhardt, R.J, Kim, Y.S, Matte, A, Cygler, M. | | Deposit date: | 2000-12-05 | | Release date: | 2001-05-02 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Active site of chondroitin AC lyase revealed by the structure of enzyme-oligosaccharide complexes and mutagenesis.
Biochemistry, 40, 2001
|
|
1HM3
 
 | | ACTIVE SITE OF CHONDROITINASE AC LYASE REVEALED BY THE STRUCTURE OF ENZYME-OLIGOSACCHARIDE COMPLEXES AND MUTAGENESIS | | Descriptor: | 2-O-methyl-beta-L-fucopyranose-(1-4)-beta-D-xylopyranose-(1-4)-alpha-D-glucopyranuronic acid-(1-2)-[alpha-L-rhamnopyranose-(1-4)]alpha-D-mannopyranose, CALCIUM ION, CHONDROITINASE AC, ... | | Authors: | Huang, W, Boju, L, Tkalec, L, Su, H, Yang, H.O, Gunay, N.S, Linhardt, R.J, Kim, Y.S, Matte, A, Cygler, M. | | Deposit date: | 2000-12-04 | | Release date: | 2001-05-02 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Active site of chondroitin AC lyase revealed by the structure of enzyme-oligosaccharide complexes and mutagenesis.
Biochemistry, 40, 2001
|
|
1HM2
 
 | | ACTIVE SITE OF CHONDROITINASE AC LYASE REVEALED BY THE STRUCTURE OF ENZYME-OLIGOSACCHARIDE COMPLEXES AND MUTAGENESIS | | Descriptor: | 2-O-methyl-beta-L-fucopyranose-(1-4)-beta-D-xylopyranose-(1-4)-alpha-D-glucopyranuronic acid-(1-2)-[alpha-L-rhamnopyranose-(1-4)]alpha-D-mannopyranose, CALCIUM ION, CHONDROITINASE AC, ... | | Authors: | Huang, W, Boju, L, Tkalec, L, Su, H, Yang, H.O, Gunay, N.S, Linhardt, R.J, Kim, Y.S, Matte, A, Cygler, M. | | Deposit date: | 2000-12-04 | | Release date: | 2001-05-02 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Active site of chondroitin AC lyase revealed by the structure of enzyme-oligosaccharide complexes and mutagenesis.
Biochemistry, 40, 2001
|
|
3DSF
 
 | | Crystal structure of anti-osteopontin antibody 23C3 in complex with W43A mutated epitope peptide | | Descriptor: | Fab fragment of anti-osteopontin antibody 23C3, Heavy chain, Light chain, ... | | Authors: | Du, J, Zhong, C, Yang, H, Ding, J. | | Deposit date: | 2008-07-12 | | Release date: | 2008-10-14 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Molecular basis of recognition of human osteopontin by 23C3, a potential therapeutic antibody for treatment of rheumatoid arthritis
J.Mol.Biol., 382, 2008
|
|
3IO8
 
 | | BimL12F in complex with Bcl-xL | | Descriptor: | Bcl-2-like protein 1, Bcl-2-like protein 11, ZINC ION | | Authors: | Colman, P.M, Lee, E.F, Fairlie, W.D, Smith, B.J, Czabotar, P.E, Yang, H, Sleebs, B.E, Lessene, G. | | Deposit date: | 2009-08-14 | | Release date: | 2009-09-01 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Conformational changes in Bcl-2 pro-survival proteins determine their capacity to bind ligands.
J.Biol.Chem., 284, 2009
|
|
6B3M
 
 | | The crystal structure of a broadly-reactive human anti-hemagglutinin stalk antibody (70-1F02) in complex with H5 hemagglutinin | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 70-1F02 Fab Heavy Chain, ... | | Authors: | Shore, D.A, Yang, H, Stevens, J. | | Deposit date: | 2017-09-22 | | Release date: | 2018-06-13 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.92 Å) | | Cite: | Broadly Reactive Human Monoclonal Antibodies Elicited following Pandemic H1N1 Influenza Virus Exposure Protect Mice against Highly Pathogenic H5N1 Challenge.
J. Virol., 92, 2018
|
|
3IO9
 
 | | BimL12Y in complex with Mcl-1 | | Descriptor: | Bcl-2-like protein 11, Induced myeloid leukemia cell differentiation protein Mcl-1, ZINC ION | | Authors: | Czabotar, P.E, Lee, E.F, Yang, H, Sleebs, B.E, Lessene, G, Colman, P.M, Smith, B.J, Fairlie, W.D. | | Deposit date: | 2009-08-14 | | Release date: | 2009-09-01 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Conformational changes in Bcl-2 pro-survival proteins determine their capacity to bind ligands.
J.Biol.Chem., 284, 2009
|
|
5QCS
 
 | | Crystal structure of BACE complex with BMC024 | | Descriptor: | (2R,4S)-N-BUTYL-4-HYDROXY-2-METHYL- 4-((E)-(4AS,12R,15S,17AS)-15-METHYL -14,17-DIOXO-2,3,4,4A,6,9,11,12,13, 14,15,16,17,17A-TETRADECAHYDRO-1H-5 ,10-DITHIA-1,13,16-TRIAZA-BENZOCYCL OPENTADECEN-12-YL)-BUTYRAMIDE, Beta-secretase 1 | | Authors: | Rondeau, J.M, Shao, C, Yang, H, Burley, S.K. | | Deposit date: | 2017-12-01 | | Release date: | 2020-06-03 | | Last modified: | 2021-02-10 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | D3R grand challenge 4: blind prediction of protein-ligand poses, affinity rankings, and relative binding free energies.
J.Comput.Aided Mol.Des., 34, 2020
|
|
5U77
 
 | | Crystal structure of ORP8 PH domain | | Descriptor: | FORMIC ACID, N-(2-hydroxyethyl)-N,N-dimethyl-3-sulfopropan-1-aminium, Oxysterol-binding protein-related protein 8 | | Authors: | Ghai, R, Yang, H. | | Deposit date: | 2016-12-11 | | Release date: | 2017-10-18 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.157 Å) | | Cite: | ORP5 and ORP8 bind phosphatidylinositol-4, 5-biphosphate (PtdIns(4,5)P 2) and regulate its level at the plasma membrane.
Nat Commun, 8, 2017
|
|
5QD4
 
 | | Crystal structure of BACE complex with BMC023 | | Descriptor: | Beta-secretase 1, {(E)-(3R,6S,9R)-3-[(1S,3R)-3-((S)-1 -BUTYLCARBAMOYL-2-METHYL-PROPYLCARB AMOYL)-1-HYDROXY-BUTYL]-6-METHYL-5, 8-DIOXO-1,11-DITHIA-4,7-DIAZA-CYCLO PENTADEC-13-EN-9-YL}-CARBAMIC ACID TERT-BUTYL ESTER | | Authors: | Rondeau, J.M, Shao, C, Yang, H, Burley, S.K. | | Deposit date: | 2017-12-01 | | Release date: | 2020-06-03 | | Last modified: | 2021-02-10 | | Method: | X-RAY DIFFRACTION (2.112 Å) | | Cite: | D3R grand challenge 4: blind prediction of protein-ligand poses, affinity rankings, and relative binding free energies.
J.Comput.Aided Mol.Des., 34, 2020
|
|
1G0H
 
 | | CRYSTAL STRUCTURE OF MJ0109 GENE PRODUCT INOSITOL MONOPHOSPHATASE-FRUCTOSE 1,6 BISPHOSPHATASE | | Descriptor: | CALCIUM ION, D-MYO-INOSITOL-1-PHOSPHATE, INOSITOL MONOPHOSPHATASE | | Authors: | Johnson, K.A, Chen, L, Yang, H, Roberts, M.F, Stec, B. | | Deposit date: | 2000-10-06 | | Release date: | 2001-03-14 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure and catalytic mechanism of the MJ0109 gene product: a bifunctional enzyme with inositol monophosphatase and fructose 1,6-bisphosphatase activities.
Biochemistry, 40, 2001
|
|
4JT6
 
 | | structure of mTORDeltaN-mLST8-PI-103 complex | | Descriptor: | 3-(4-MORPHOLIN-4-YLPYRIDO[3',2':4,5]FURO[3,2-D]PYRIMIDIN-2-YL)PHENOL, mLST8, mTOR | | Authors: | Pavletich, N.P, Yang, H. | | Deposit date: | 2013-03-22 | | Release date: | 2013-05-08 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | mTOR kinase structure, mechanism and regulation.
Nature, 497, 2013
|
|
1G0I
 
 | | CRYSTAL STRUCTURE OF MJ0109 GENE PRODUCT INOSITOL MONOPHOSPHATASE-FRUCTOSE 1,6 BISPHOSPHATASE | | Descriptor: | 1,2,3,4,5,6-HEXAHYDROXY-CYCLOHEXANE, INOSITOL MONOPHOSPHATASE, MANGANESE (II) ION, ... | | Authors: | Johnson, K.A, Chen, L, Yang, H, Roberts, M.F, Stec, B. | | Deposit date: | 2000-10-06 | | Release date: | 2001-03-14 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure and catalytic mechanism of the MJ0109 gene product: a bifunctional enzyme with inositol monophosphatase and fructose 1,6-bisphosphatase activities.
Biochemistry, 40, 2001
|
|
