5DKZ
 
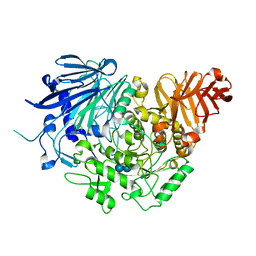 | | Crystal structure of glucosidase II alpha subunit (alpha3-Glc2-bound from) | | Descriptor: | Alpha glucosidase-like protein, alpha-D-glucopyranose-(1-3)-alpha-D-glucopyranose | | Authors: | Satoh, T, Toshimori, T, Yan, G, Yamaguchi, T, Kato, K. | | Deposit date: | 2015-09-04 | | Release date: | 2016-01-27 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis for two-step glucose trimming by glucosidase II involved in ER glycoprotein quality control.
Sci Rep, 6, 2016
|
|
5DKX
 
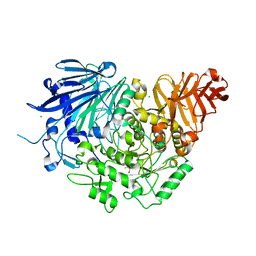 | | Crystal structure of glucosidase II alpha subunit (Tris-bound from) | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Alpha glucosidase-like protein, CHLORIDE ION | | Authors: | Satoh, T, Toshimori, T, Yan, G, Yamaguchi, T, Kato, K. | | Deposit date: | 2015-09-04 | | Release date: | 2016-01-27 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural basis for two-step glucose trimming by glucosidase II involved in ER glycoprotein quality control.
Sci Rep, 6, 2016
|
|
5DL0
 
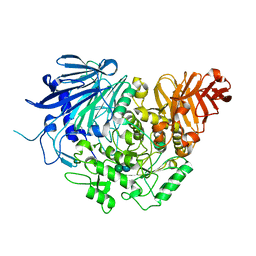 | | Crystal structure of glucosidase II alpha subunit (Glc1Man2-bound from) | | Descriptor: | Alpha glucosidase-like protein, alpha-D-glucopyranose-(1-3)-alpha-D-mannopyranose | | Authors: | Satoh, T, Toshimori, T, Yan, G, Yamaguchi, T, Kato, K. | | Deposit date: | 2015-09-04 | | Release date: | 2016-01-27 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis for two-step glucose trimming by glucosidase II involved in ER glycoprotein quality control.
Sci Rep, 6, 2016
|
|
4R12
 
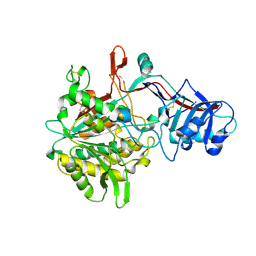 | | Crystal structure of the gamma-secretase component Nicastrin | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Xie, T, Yan, C, Zhou, R, Zhao, Y, Sun, L, Yang, G, Lu, P, Ma, D, Shi, Y. | | Deposit date: | 2014-08-03 | | Release date: | 2014-09-17 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structure of the gamma-secretase component nicastrin.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
5A63
 
 | | Cryo-EM structure of the human gamma-secretase complex at 3.4 angstrom resolution. | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Bai, X, Yan, C, Yang, G, Lu, P, Ma, D, Sun, L, Zhou, R, Scheres, S.H.W, Shi, Y. | | Deposit date: | 2015-06-24 | | Release date: | 2015-08-05 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | An Atomic Structure of Human Gamma-Secretase
Nature, 525, 2015
|
|
5XLS
 
 | | Crystal structure of UraA in occluded conformation | | Descriptor: | 12-TUNGSTOPHOSPHATE, URACIL, Uracil permease | | Authors: | Yu, X.Z, Yang, G.H, Yan, C.Y, Yan, N. | | Deposit date: | 2017-05-11 | | Release date: | 2017-07-05 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Dimeric structure of the uracil:proton symporter UraA provides mechanistic insights into the SLC4/23/26 transporters
Cell Res., 27, 2017
|
|
6L7I
 
 | |
6L7E
 
 | |
8X7Z
 
 | |
7YD3
 
 | | Single-chain variable fragment of app 3D1 antibody | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Yan, L, Yang, G. | | Deposit date: | 2022-07-03 | | Release date: | 2023-07-05 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Cross-reactive epitopes between HIV and Coronavirus revealed by 3D1
To Be Published
|
|
7YI6
 
 | |
5DKY
 
 | | Crystal structure of glucosidase II alpha subunit (DNJ-bound from) | | Descriptor: | 1-DEOXYNOJIRIMYCIN, Alpha glucosidase-like protein | | Authors: | Satoh, T, Toshimori, T, Yan, G, Yamaguchi, T, Kato, K. | | Deposit date: | 2015-09-04 | | Release date: | 2016-01-27 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural basis for two-step glucose trimming by glucosidase II involved in ER glycoprotein quality control.
Sci Rep, 6, 2016
|
|
6JX9
 
 | | Structure of Y17107 complexed HPPD | | Descriptor: | 4-hydroxyphenylpyruvate dioxygenase, 5-[(~{Z})-(1,3-dimethyl-5-oxidanylidene-pyrazol-4-ylidene)-oxidanyl-methyl]-2-(phenylmethyl)isoindole-1,3-dione, COBALT (II) ION | | Authors: | Lin, H.Y, Yang, G.F. | | Deposit date: | 2019-04-22 | | Release date: | 2020-03-11 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Pyrazole-Isoindoline-1,3-dione Hybrid: A Promising Scaffold for 4-Hydroxyphenylpyruvate Dioxygenase Inhibitors.
J.Agric.Food Chem., 67, 2019
|
|
2ADU
 
 | | Human Methionine Aminopeptidase Complex with 4-Aryl-1,2,3-triazole Inhibitor | | Descriptor: | 4-(3-METHYLPHENYL)-1H-1,2,3-TRIAZOLE, COBALT (II) ION, Methionine aminopeptidase 2 | | Authors: | Kallander, L.S, Lu, Q, Chen, W, Tomaszek, T, Yang, G, Tew, D, Meek, T.D, Hofmann, G.A, Schulz-Pritchard, C.K, Smith, W.W, Janson, C.A, Ryan, M.D, Zhang, G.F, Johanson, K.O, Kirkpatrick, R.B, Ho, T.F, Fisher, P.W, Mattern, M.R, Johnson, R.K, Hansbury, M.J, Winkler, J.D, Ward, K.W, Veber, D.F, Thompson, S.K. | | Deposit date: | 2005-07-20 | | Release date: | 2005-09-13 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | 4-Aryl-1,2,3-triazole: A Novel Template for a Reversible Methionine Aminopeptidase 2 Inhibitor, Optimized To Inhibit Angiogenesis in Vivo
J.Med.Chem., 48, 2005
|
|
2OAZ
 
 | | Human Methionine Aminopeptidase-2 Complexed with SB-587094 | | Descriptor: | COBALT (II) ION, N-(2-ISOPROPYLPHENYL)-3-[(2-THIENYLMETHYL)THIO]-1H-1,2,4-TRIAZOL-5-AMINE, human Methionine Amino Peptidase 2 | | Authors: | Marino Jr, J.P, Fisher, P.W, Hofmann, G.A, Kirkpatrick, R, Janson, C.A, Johnson, R.K, Ma, C, Mattern, M, Meek, T.D, Ryan, D, Schulz, C, Smith, W.W, Tew, D.G, Tomazek Jr, T.A, Veber, D.F, Xiong, W.C, Yamamoto, Y, Yamashita, K, Yang, G, Thompson, S.K. | | Deposit date: | 2006-12-18 | | Release date: | 2007-06-19 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Highly potent inhibitors of methionine aminopeptidase-2 based on a 1,2,4-triazole pharmacophore.
J.Med.Chem., 50, 2007
|
|
4U3F
 
 | | Cytochrome bc1 complex from chicken with designed inhibitor bound | | Descriptor: | 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CARDIOLIPIN, ... | | Authors: | Huang, L.-S, Zhu, X.-L, Yang, G.F, Berry, E.A. | | Deposit date: | 2014-07-21 | | Release date: | 2015-07-29 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.2312 Å) | | Cite: | Rational Design of Highly Potent and Slow-Binding Cytochrome bc1 Inhibitor as Fungicide by Computational Substitution Optimization
Sci Rep, 5, 2015
|
|
9CV6
 
 | |
3TGU
 
 | | Cytochrome bc1 complex from chicken with pfvs-designed moa inhibitor bound | | Descriptor: | 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, CARDIOLIPIN, Coenzyme Q10, ... | | Authors: | Huang, L.-S, Yang, G.-F, Berry, E.A. | | Deposit date: | 2011-08-17 | | Release date: | 2012-07-04 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Computational discovery of picomolar Q(o) site inhibitors of cytochrome bc1 complex.
J.Am.Chem.Soc., 134, 2012
|
|
4WVX
 
 | | Crystal structure of a phosphotriesterase-like lactonase Gkap in native form | | Descriptor: | 1,2-ETHANEDIOL, COBALT (II) ION, Phosphotriesterase | | Authors: | An, J, Zhang, Y, Yang, G.Y, Feng, Y. | | Deposit date: | 2014-11-07 | | Release date: | 2015-11-11 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Engineering a thermostable lactonase for enhanced phosphotriesterase activity against organophosphate pesticides
To Be Published
|
|
1JXZ
 
 | | Structure of the H90Q mutant of 4-Chlorobenzoyl-Coenzyme A Dehalogenase complexed with 4-hydroxybenzoyl-Coenzyme A (product) | | Descriptor: | 4-HYDROXYBENZOYL COENZYME A, 4-chlorobenzoyl Coenzyme A dehalogenase, CALCIUM ION, ... | | Authors: | Thoden, J.B, Zhang, W, Wei, Y, Luo, L, Taylor, K.L, Yang, G, Dunaway-Mariano, D, Benning, M.M, Holden, H.M. | | Deposit date: | 2001-09-10 | | Release date: | 2001-10-03 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Histidine 90 Function in 4-chlorobenzoyl-coenzyme A Dehalogenase Catalysis
Biochemistry, 40, 2001
|
|
3G5K
 
 | | Structure and activity of human mitochondrial peptide deformylase, a novel cancer target | | Descriptor: | ACTINONIN, COBALT (II) ION, Peptide deformylase, ... | | Authors: | Escobar-Alvarez, S, Goldgur, Y, Yang, G, Ouerfelli, O, Li, Y, Scheinberg, D.A. | | Deposit date: | 2009-02-05 | | Release date: | 2009-04-07 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure and activity of human mitochondrial peptide deformylase, a novel cancer target
J.Mol.Biol., 387, 2009
|
|
3G5P
 
 | | Structure and activity of human mitochondrial peptide deformylase, a novel cancer target | | Descriptor: | COBALT (II) ION, PHOSPHATE ION, Peptide deformylase, ... | | Authors: | Escobar-Alvarez, S, Goldgur, Y, Yang, G, Ouerfelli, O, Li, Y, Scheinberg, D.A. | | Deposit date: | 2009-02-05 | | Release date: | 2009-04-07 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure and activity of human mitochondrial peptide deformylase, a novel cancer target
J.Mol.Biol., 387, 2009
|
|
6ISD
 
 | | Crystal structure of Arabidopsis thaliana HPPD complexed with sulcotrione | | Descriptor: | 2-[2-chloro-4-(methylsulfonyl)benzoyl]cyclohexane-1,3-dione, 4-hydroxyphenylpyruvate dioxygenase, COBALT (II) ION | | Authors: | Yang, W.C, Yang, G.F. | | Deposit date: | 2018-11-16 | | Release date: | 2018-11-28 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Molecular insights into the mechanism of 4-hydroxyphenylpyruvate dioxygenase inhibition: enzyme kinetics, X-ray crystallography and computational simulations.
FEBS J., 286, 2019
|
|
6J63
 
 | | Crystal structure of Arabidopsis thaliana HPPD complexed with NTBC | | Descriptor: | 2-{HYDROXY[2-NITRO-4-(TRIFLUOROMETHYL)PHENYL]METHYLENE}CYCLOHEXANE-1,3-DIONE, 4-hydroxyphenylpyruvate dioxygenase, FE (III) ION | | Authors: | Yang, W.C, Yang, G.F. | | Deposit date: | 2019-01-13 | | Release date: | 2019-02-06 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.624 Å) | | Cite: | Molecular insights into the mechanism of 4-hydroxyphenylpyruvate dioxygenase inhibition: enzyme kinetics, X-ray crystallography and computational simulations.
FEBS J., 286, 2019
|
|
3UIV
 
 | | Human serum albumin-myristate-amantadine hydrochloride complex | | Descriptor: | (3S,5S,7S)-tricyclo[3.3.1.1~3,7~]decan-1-amine, MYRISTIC ACID, Serum albumin | | Authors: | Yang, F, Ma, Z, Ma, L, Yang, G. | | Deposit date: | 2011-11-06 | | Release date: | 2012-11-07 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis of the drug-binding specificity of human serum albumin
TO BE PUBLISHED
|
|
