3W8E
 
 | | Crystal structure of D-3-hydroxybutyrate dehydrogenase from Alcaligenes faecalis complexed with NAD+ and a substrate D-3-hydroxybutyrate | | Descriptor: | (3R)-3-hydroxybutanoic acid, D-3-hydroxybutyrate dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Kanazawa, H, Tsunoda, M, Hoque, M.M, Suzuki, K, Yamamoto, T, Takenaka, A. | | Deposit date: | 2013-03-12 | | Release date: | 2014-03-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.24 Å) | | Cite: | High resolution X-ray diffraction of D-3-hydroxybutyrate dehydrogenase from Alcaligenes faecalis complexed with NAD+ and D-3-hydroxybutyrate
To be Published
|
|
3W8F
 
 | | Crystal structure of D-3-hydroxybutyrate dehydrogenase from Alcaligenes faecalis complexed with NAD+ and an inhibitor malonate | | Descriptor: | CHLORIDE ION, D-3-hydroxybutyrate dehydrogenase, MALONIC ACID, ... | | Authors: | Kanazawa, H, Tsunoda, M, Hoque, M.M, Suzuki, K, Yamamoto, T, Takenaka, A. | | Deposit date: | 2013-03-12 | | Release date: | 2014-03-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | X-ray diffraction of D-3-hydroxybutyrate dehydrogenase from Alcaligenes faecalis complexed with NAD+ and malonate
To be Published
|
|
6JT4
 
 | | Crystal Structure of BACE1 in complex with N-{3-[(4S,6S)-2-amino-4-methyl-6-(trifluoromethyl)-5,6-dihydro-4H-1,3-thiazin-4-yl]-4-fluorophenyl}-5-(fluoromethoxy)pyrazine-2-carboxamide | | Descriptor: | Beta-secretase 1, GLYCEROL, IODIDE ION, ... | | Authors: | Anan, K, Iso, Y, Oguma, T, Nakahara, K, Suzuki, S, Yamamoto, T, Matsuoka, E, Ito, H, Sakaguchi, G, Ando, S, Morimoto, K, Kanegawa, N, Kido, Y, Kawachi, T, Fukushima, T, Teisman, A, Urmaliya, V, Dhuyvetter, D, Borghys, H, Austin, N, Bergh, A.V.D, Verboven, P, Bischoff, F, Gijsen, H.J.M, Yamano, Y, Kusakabe, K.I. | | Deposit date: | 2019-04-08 | | Release date: | 2020-01-15 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Trifluoromethyl Dihydrothiazine-Based beta-Secretase (BACE1) Inhibitors with Robust Central beta-Amyloid Reduction and Minimal Covalent Binding Burden.
Chemmedchem, 14, 2019
|
|
6JT3
 
 | | Crystal Structure of BACE1 in complex with N-{3-[(4R,5R,6R)-2-amino-5-fluoro-4,6-dimethyl-5,6-dihydro-4H-1,3-thiazin-4-yl]-4-fluorophenyl}-5-(fluoromethoxy)pyrazine-2-carboxamide | | Descriptor: | Beta-secretase 1, DIMETHYL SULFOXIDE, GLYCEROL, ... | | Authors: | Tadano, G, Komano, K, Yoshida, S, Suzuki, S, Nakahara, K, Fuchino, K, Fujimoto, K, Matsuoka, E, Yamamoto, T, Asada, N, Ito, H, Sakaguchi, G, Kanegawa, N, Kido, Y, Ando, S, Fukushima, T, Teisman, A, Urmaliya, V, Dhuyvetter, D, Borghys, H, Bergh, A.V.D, Austin, N, Gijsen, H.J.M, Yamano, Y, Iso, Y, Kusakabe, K.I. | | Deposit date: | 2019-04-08 | | Release date: | 2019-10-30 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Discovery of an Extremely Potent Thiazine-Based beta-Secretase Inhibitor with Reduced Cardiovascular and Liver Toxicity at a Low Projected Human Dose.
J.Med.Chem., 62, 2019
|
|
2E2G
 
 | | Crystal structure of archaeal peroxiredoxin, thioredoxin peroxidase from Aeropyrum pernix K1 (pre-oxidation form) | | Descriptor: | Probable peroxiredoxin | | Authors: | Nakamura, T, Yamamoto, T, Abe, M, Matsumura, H, Hagihara, Y, Goto, T, Yamaguchi, T, Inoue, T. | | Deposit date: | 2006-11-13 | | Release date: | 2007-11-20 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Oxidation of archaeal peroxiredoxin involves a hypervalent sulfur intermediate
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
2EB2
 
 | | Crystal structure of mutated EGFR kinase domain (G719S) | | Descriptor: | Epidermal growth factor receptor | | Authors: | Yoshikawa, S, Kukimoto-Niino, M, Chen, L, Liu, Z.J, Wang, B.C, Shirouzu, M, Senba, K, Yamamoto, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-02-06 | | Release date: | 2008-02-12 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis for the altered drug sensitivities of non-small cell lung cancer-associated mutants of human epidermal growth factor receptor
Oncogene, 2012
|
|
2E2M
 
 | | Crystal structure of archaeal peroxiredoxin, thioredoxin peroxidase from Aeropyrum pernix K1 (sulfinic acid form) | | Descriptor: | Probable peroxiredoxin | | Authors: | Nakamura, T, Yamamoto, T, Abe, M, Matsumura, H, Hagihara, Y, Goto, T, Yamaguchi, T, Inoue, T. | | Deposit date: | 2006-11-14 | | Release date: | 2007-11-20 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Oxidation of archaeal peroxiredoxin involves a hypervalent sulfur intermediate
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
6JG0
 
 | | Crystal structure of the N-terminal domain single mutant (S92E) of the human mitochondrial calcium uniporter fused with T4 lysozyme | | Descriptor: | Endolysin,Calcium uniporter protein, SULFATE ION | | Authors: | Lee, Y, Park, J, Min, C.K, Kang, J.Y, Kim, T.G, Yamamoto, T, Kim, D.H, Eom, S.H. | | Deposit date: | 2019-02-13 | | Release date: | 2020-02-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of calcium channel domain
To Be Published
|
|
2EB3
 
 | | Crystal structure of mutated EGFR kinase domain (L858R) in complex with AMPPNP | | Descriptor: | Epidermal growth factor receptor, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER | | Authors: | Yoshikawa, S, Kukimoto-Niino, M, Shirouzu, M, Senba, K, Yamamoto, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-02-06 | | Release date: | 2008-02-12 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.84 Å) | | Cite: | Structural basis for the altered drug sensitivities of non-small cell lung cancer-associated mutants of human epidermal growth factor receptor
Oncogene, 2012
|
|
2D5R
 
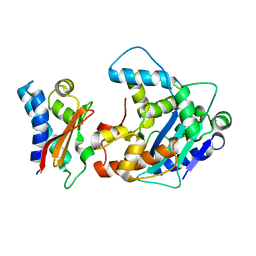 | | Crystal Structure of a Tob-hCaf1 Complex | | Descriptor: | CCR4-NOT transcription complex subunit 7, Tob1 protein | | Authors: | Horiuchi, M, Suzuki, N.N, Muroya, N, Takahasi, K, Nishida, M, Yoshida, Y, Ikematsu, N, Nakamura, T, Kawamura-Tsuzuku, J, Yamamoto, T, Inagaki, F. | | Deposit date: | 2005-11-04 | | Release date: | 2006-12-12 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis for the antiproliferative activity of the Tob-hCaf1 complex.
J.Biol.Chem., 284, 2009
|
|
2DJI
 
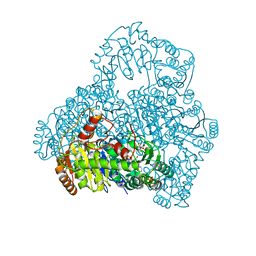 | | Crystal Structure of Pyruvate Oxidase from Aerococcus viridans containing FAD | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Pyruvate oxidase, SULFATE ION | | Authors: | Juan, E.C.M, Hossain, M.T, Suzuki, K, Yamamoto, T, Imamura, S, Sekiguchi, T, Takenaka, A. | | Deposit date: | 2006-04-03 | | Release date: | 2007-05-29 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The structures of pyruvate oxidase from Aerococcus viridans with cofactors and with a reaction intermediate reveal the flexibility of the active-site tunnel for catalysis.
Acta Crystallogr.,Sect.F, 63, 2007
|
|
2DV6
 
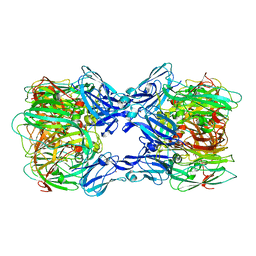 | | Crystal structure of nitrite reductase from Hyphomicrobium denitrificans | | Descriptor: | COPPER (II) ION, Nitrite reductase, POTASSIUM ION | | Authors: | Nojiri, M, Xie, Y, Yamamoto, T, Inoue, T, Suzuki, S, Kai, Y. | | Deposit date: | 2006-07-28 | | Release date: | 2007-02-20 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure and function of a hexameric copper-containing nitrite reductase.
Proc.Natl.Acad.Sci.USA, 104, 2007
|
|
6JSG
 
 | | Crystal Structure of BACE1 in complex with N-{3-[(4S)-2-amino-4-methyl-5,6-dihydro-4H-1,3-thiazin-4-yl]-4-fluorophenyl}-5-chloropyridine-2-carboxamide | | Descriptor: | Beta-secretase 1, GLYCEROL, IODIDE ION, ... | | Authors: | Fujimoto, K, Matsuoka, E, Asada, N, Tadano, G, Yamamoto, T, Nakahara, K, Fuchino, K, Ito, H, Kanegawa, N, Moechars, D, Gijsen, H.J.M, Kusakabe, K.I. | | Deposit date: | 2019-04-08 | | Release date: | 2019-08-28 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure-Based Design of Selective beta-Site Amyloid Precursor Protein Cleaving Enzyme 1 (BACE1) Inhibitors: Targeting the Flap to Gain Selectivity over BACE2.
J.Med.Chem., 62, 2019
|
|
6JSF
 
 | | Crystal Structure of BACE1 in complex with N-(3-((4S,5S)-2-amino-4-methyl-5-phenyl-5,6-dihydro-4H-1,3-thiazin-4-yl)-4-fluorophenyl)-5-(fluoromethoxy)pyrazine-2-carboxamide | | Descriptor: | Beta-secretase 1, DIMETHYL SULFOXIDE, IODIDE ION, ... | | Authors: | Fujimoto, K, Matsuoka, E, Asada, N, Tadano, G, Yamamoto, T, Nakahara, K, Fuchino, K, Ito, H, Kanegawa, N, Moechars, D, Gijsen, H.J.M, Kusakabe, K.I. | | Deposit date: | 2019-04-08 | | Release date: | 2019-08-28 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure-Based Design of Selective beta-Site Amyloid Precursor Protein Cleaving Enzyme 1 (BACE1) Inhibitors: Targeting the Flap to Gain Selectivity over BACE2.
J.Med.Chem., 62, 2019
|
|
6JSZ
 
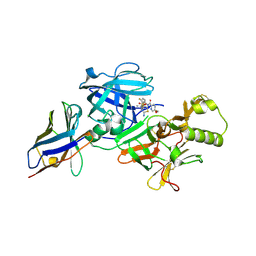 | | BACE2 xaperone complex with N-{3-[(5R)-3-amino-5-methyl-9,9-dioxo-2,9lambda6-dithia-4-azaspiro[5.5]undec-3-en-5-yl]-4-fluorophenyl}-5-(fluoromethoxy)pyrazine-2-carboxamide | | Descriptor: | Beta-secretase 2, CHLORIDE ION, N-[3-[(5R)-3-azanyl-5-methyl-9,9-bis(oxidanylidene)-2,9$l^{6}-dithia-4-azaspiro[5.5]undec-3-en-5-yl]-4-fluoranyl-phenyl]-5-(fluoranylmethoxy)pyrazine-2-carboxamide, ... | | Authors: | Fujimoto, K, Matsuoka, E, Asada, N, Tadano, G, Yamamoto, T, Nakahara, K, Fuchino, K, Ito, H, Kanegawa, N, Moechars, D, Gijsen, H.J.M, Kusakabe, K.I. | | Deposit date: | 2019-04-08 | | Release date: | 2019-08-28 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Structure-Based Design of Selective beta-Site Amyloid Precursor Protein Cleaving Enzyme 1 (BACE1) Inhibitors: Targeting the Flap to Gain Selectivity over BACE2.
J.Med.Chem., 62, 2019
|
|
6JSN
 
 | | Crystal Structure of BACE1 in complex with N-{3-[(5R)-3-amino-5-methyl-9,9-dioxo-2,9lambda6-dithia-4-azaspiro[5.5]undec-3-en-5-yl]-4-fluorophenyl}-5-(fluoromethoxy)pyrazine-2-carboxamide | | Descriptor: | Beta-secretase 1, GLYCEROL, IODIDE ION, ... | | Authors: | Fujimoto, K, Matsuoka, E, Asada, N, Tadano, G, Yamamoto, T, Nakahara, K, Fuchino, K, Ito, H, Kanegawa, N, Moechars, D, Gijsen, H.J.M, Kusakabe, K.I. | | Deposit date: | 2019-04-08 | | Release date: | 2019-08-28 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure-Based Design of Selective beta-Site Amyloid Precursor Protein Cleaving Enzyme 1 (BACE1) Inhibitors: Targeting the Flap to Gain Selectivity over BACE2.
J.Med.Chem., 62, 2019
|
|
6JSE
 
 | | Crystal Structure of BACE1 in complex with N-(3-((4S,5R)-2-amino-4-methyl-5-phenyl-5,6-dihydro-4H-1,3-thiazin-4-yl)-4-fluorophenyl)-5-(fluoromethoxy)pyrazine-2-carboxamide | | Descriptor: | Beta-secretase 1, DIMETHYL SULFOXIDE, IODIDE ION, ... | | Authors: | Fujimoto, K, Matsuoka, E, Asada, N, Tadano, G, Yamamoto, T, Nakahara, K, Fuchino, K, Ito, H, Kanegawa, N, Moechars, D, Gijsen, H.J.M, Kusakabe, K.I. | | Deposit date: | 2019-04-08 | | Release date: | 2019-08-28 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure-Based Design of Selective beta-Site Amyloid Precursor Protein Cleaving Enzyme 1 (BACE1) Inhibitors: Targeting the Flap to Gain Selectivity over BACE2.
J.Med.Chem., 62, 2019
|
|
5AXE
 
 | | Crystal Structure Analysis of DNA Duplexes containing sulfoamide-bridged nucleic acid (SuNA-NH) | | Descriptor: | DNA (5'-D(*GP*CP*GP*TP*AP*(LSH)P*AP*CP*GP*C)-3') | | Authors: | Mitsuoka, Y, Aoyama, H, Kugimiya, A, Fujimura, Y, Yamamoto, T, Waki, R, Wada, F, Tahara, S, Sawamura, M, Noda, M, Hari, Y, Obika, S. | | Deposit date: | 2015-07-28 | | Release date: | 2016-07-20 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (0.95 Å) | | Cite: | Effect of an N-substituent in sulfonamide-bridged nucleic acid (SuNA) on hybridization ability and duplex structure.
Org.Biomol.Chem., 14, 2016
|
|
5AXF
 
 | | Crystal Structure Analysis of DNA Duplexes containing sulfoamide-bridged nucleic acid (SuNA-NMe) | | Descriptor: | DNA (5'-D(*GP*CP*GP*TP*AP*(LSM)P*AP*CP*GP*C)-3') | | Authors: | Mitsuoka, Y, Aoyama, H, Kugimiya, A, Fujimura, Y, Yamamoto, T, Waki, R, Wada, F, Tahara, S, Sawamura, M, Noda, M, Hari, Y, Obika, S. | | Deposit date: | 2015-07-28 | | Release date: | 2016-07-20 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.13 Å) | | Cite: | Effect of an N-substituent in sulfonamide-bridged nucleic acid (SuNA) on hybridization ability and duplex structure.
Org.Biomol.Chem., 14, 2016
|
|
5B4V
 
 | | Crystal structure of D-3-hydroxybutyrate dehydrogenase from Alcaligenes faecalis complexed with NAD+ and an inhibitor methylmalonate | | Descriptor: | 3-hydroxybutyrate dehydrogenase, CHLORIDE ION, METHYLMALONIC ACID, ... | | Authors: | Kanazawa, H, Tsunoda, M, Hoque, M.M, Suzuki, K, Yamamoto, T, Takenaka, A. | | Deposit date: | 2016-04-19 | | Release date: | 2016-08-17 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural insights into the catalytic reaction trigger and inhibition of D-3-hydroxybutyrate dehydrogenase
Acta Crystallogr.,Sect.F, 72, 2016
|
|
5B4T
 
 | | Crystal structure of D-3-hydroxybutyrate dehydrogenase from Alcaligenes faecalis complexed with NAD+ and a substrate D-3-hydroxybutyrate | | Descriptor: | (3R)-3-hydroxybutanoic acid, 3-hydroxybutyrate dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Kanazawa, H, Tsunoda, M, Hoque, M.M, Suzuki, K, Yamamoto, T, Takenaka, A. | | Deposit date: | 2016-04-19 | | Release date: | 2016-08-17 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.19 Å) | | Cite: | Structural insights into the catalytic reaction trigger and inhibition of D-3-hydroxybutyrate dehydrogenase
Acta Crystallogr.,Sect.F, 72, 2016
|
|
5B4U
 
 | | Crystal structure of D-3-hydroxybutyrate dehydrogenase from Alcaligenes faecalis complexed with NAD+ and an inhibitor malonate | | Descriptor: | 3-hydroxybutyrate dehydrogenase, CHLORIDE ION, MALONIC ACID, ... | | Authors: | Kanazawa, H, Tsunoda, M, Hoque, M.M, Suzuki, K, Yamamoto, T, Takenaka, A. | | Deposit date: | 2016-04-19 | | Release date: | 2016-08-17 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structural insights into the catalytic reaction trigger and inhibition of D-3-hydroxybutyrate dehydrogenase
Acta Crystallogr.,Sect.F, 72, 2016
|
|
6KVX
 
 | | Crystal structure of the N-terminal domain single mutant (D119A) of the human mitochondrial calcium uniporter fused with T4 lysozyme | | Descriptor: | Endolysin,Calcium uniporter protein, mitochondrial, SULFATE ION | | Authors: | Lee, Y, Park, J, Lee, G, Yoon, S, Min, C.K, Kim, T.G, Yamamoto, T, Kim, D.H, Lee, K.W, Eom, S.H. | | Deposit date: | 2019-09-05 | | Release date: | 2020-05-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Crystal structure of the N-terminal domain single mutant (D119A) of the human mitochondrial calcium uniporter fused with T4 lysozyme
Sci Rep, 2020
|
|
