8IW1
 
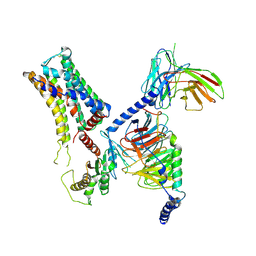 | | Cryo-EM structure of the PEA-bound mTAAR9-Golf complex | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(i) subunit alpha-1,Guanine nucleotide-binding protein G(olf) subunit alpha, ... | | Authors: | Sun, J.P, Li, Q, Yang, F, Xu, Y.F, Guo, L.L, Lian, S, Zhang, M.H, Rong, N.K. | | Deposit date: | 2023-03-29 | | Release date: | 2023-05-31 | | Last modified: | 2023-06-14 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural basis of amine odorant perception by a mammal olfactory receptor.
Nature, 618, 2023
|
|
8IW4
 
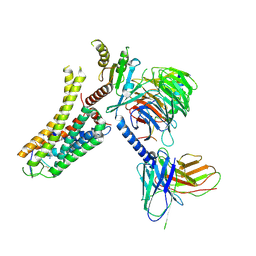 | | Cryo-EM structure of the SPE-bound mTAAR9-Gs complex | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(s) subunit alpha isoforms short, ... | | Authors: | Sun, J.P, Li, Q, Yang, F, Xu, Y.F, Guo, L.L, Lian, S, Zhang, M.H, Rong, N.K. | | Deposit date: | 2023-03-29 | | Release date: | 2023-05-31 | | Last modified: | 2023-06-14 | | Method: | ELECTRON MICROSCOPY (3.49 Å) | | Cite: | Structural basis of amine odorant perception by a mammal olfactory receptor.
Nature, 618, 2023
|
|
8ITF
 
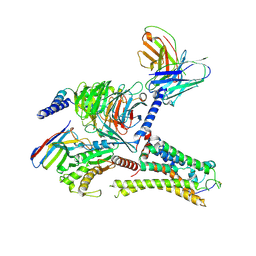 | | Cryo-EM structure of the DMCHA-bound mTAAR9-Gs complex | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(s) subunit alpha isoforms short, ... | | Authors: | Sun, J.P, Li, Q, Yang, F, Xu, Y.F, Guo, L.L, Lian, S, Zhang, M.H, Rong, N.K. | | Deposit date: | 2023-03-22 | | Release date: | 2023-05-31 | | Last modified: | 2023-06-14 | | Method: | ELECTRON MICROSCOPY (3.46 Å) | | Cite: | Structural basis of amine odorant perception by a mammal olfactory receptor.
Nature, 618, 2023
|
|
8IW7
 
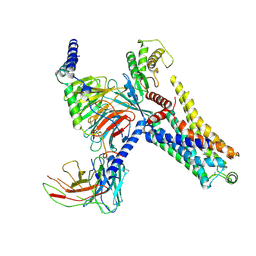 | | Cryo-EM structure of the PEA-bound mTAAR9-Gs complex | | Descriptor: | 2-PHENYLETHYLAMINE, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Sun, J.P, Li, Q, Yang, F, Xu, Y.F, Guo, L.L, Lian, S, Zhang, M.H, Rong, N.K. | | Deposit date: | 2023-03-29 | | Release date: | 2023-05-31 | | Last modified: | 2023-06-14 | | Method: | ELECTRON MICROSCOPY (2.97 Å) | | Cite: | Structural basis of amine odorant perception by a mammal olfactory receptor.
Nature, 618, 2023
|
|
8IW9
 
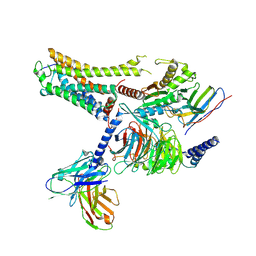 | | Cryo-EM structure of the CAD-bound mTAAR9-Gs complex | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(s) subunit alpha isoforms short, ... | | Authors: | Sun, J.P, Li, Q, Yang, F, Xu, Y.F, Guo, L.L, Lian, S, Zhang, M.H, Rong, N.K. | | Deposit date: | 2023-03-29 | | Release date: | 2023-05-31 | | Last modified: | 2023-06-14 | | Method: | ELECTRON MICROSCOPY (3.08 Å) | | Cite: | Structural basis of amine odorant perception by a mammal olfactory receptor.
Nature, 618, 2023
|
|
8IWM
 
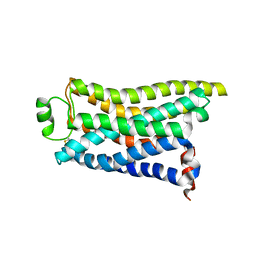 | | Cryo-EM structure of the PEA-bound mTAAR9 complex | | Descriptor: | 2-PHENYLETHYLAMINE, Trace amine-associated receptor 9 | | Authors: | Sun, J.P, Li, Q, Yang, F, Xu, Y.F, Guo, L.L, Lian, S, Zhang, M.H, Rong, N.K. | | Deposit date: | 2023-03-30 | | Release date: | 2023-05-31 | | Last modified: | 2023-06-14 | | Method: | ELECTRON MICROSCOPY (3.17 Å) | | Cite: | Structural basis of amine odorant perception by a mammal olfactory receptor.
Nature, 618, 2023
|
|
8W8F
 
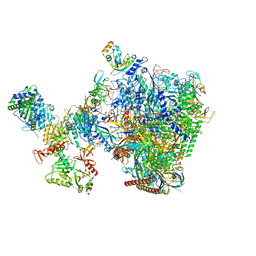 | | human co-transcriptional RNA capping enzyme RNGTT-CMTR1 | | Descriptor: | Cap-specific mRNA (nucleoside-2'-O-)-methyltransferase 1, DNA (36-MER), DNA (45-MER), ... | | Authors: | Li, Y, Wang, Q, Xu, Y, Li, Z. | | Deposit date: | 2023-09-02 | | Release date: | 2024-04-10 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Strcutures of co-transcriptional RNA capping enzymes on paused transcription complex
To Be Published
|
|
8W8E
 
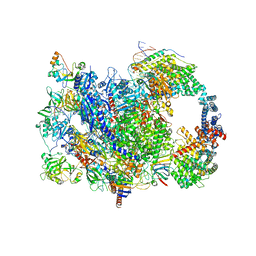 | | human co-transcriptional RNA capping enzyme RNGTT | | Descriptor: | DNA (36-MER), DNA (45-MER), DNA-directed RNA polymerase II subunit E, ... | | Authors: | Li, Y, Wang, Q, Xu, Y, Li, Z. | | Deposit date: | 2023-09-02 | | Release date: | 2024-04-10 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Strcutures of co-transcriptional RNA capping enzymes on paused transcription complex
To Be Published
|
|
8Y33
 
 | | A near-infrared fluorescent protein of de novo backbone design | | Descriptor: | 3-[5-[(4-ethenyl-3-methyl-5-oxidanylidene-pyrrol-2-yl)methyl]-2-[[5-[(3-ethyl-4-methyl-5-oxidanylidene-pyrrol-2-yl)methyl]-3-(3-hydroxy-3-oxopropyl)-4-methyl-1~{H}-pyrrol-2-yl]methyl]-4-methyl-1~{H}-pyrrol-3-yl]propanoic acid, near-infrared fluorescent protein | | Authors: | Hu, X, Xu, Y. | | Deposit date: | 2024-01-28 | | Release date: | 2024-02-28 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Using Protein Design and Directed Evolution to Monomerize a Bright Near-Infrared Fluorescent Protein.
Acs Synth Biol, 13, 2024
|
|
6LPC
 
 | | Crystal Structure of rat Munc18-1 with K332E/K333E mutation | | Descriptor: | Syntaxin-binding protein 1 | | Authors: | Wang, X.P, Gong, J.H, Wang, S, Zhu, L, Yang, X.Y, Xu, Y.Y, Yang, X.F, Ma, C. | | Deposit date: | 2020-01-09 | | Release date: | 2020-07-15 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.402 Å) | | Cite: | Munc13 activates the Munc18-1/syntaxin-1 complex and enables Munc18-1 to prime SNARE assembly.
Embo J., 39, 2020
|
|
8HI9
 
 | | SARS-CoV-2 3CL protease (3CLpro) in complex with Robinetin | | Descriptor: | 3,7-bis(oxidanyl)-2-[3,4,5-tris(oxidanyl)phenyl]chromen-4-one, 3C-like proteinase nsp5 | | Authors: | Su, H.X, Xie, H, Li, M.J, Xu, Y.C. | | Deposit date: | 2022-11-19 | | Release date: | 2023-10-25 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Discovery of Polyphenolic Natural Products as SARS-CoV-2 M pro Inhibitors for COVID-19.
Pharmaceuticals, 16, 2023
|
|
6LJS
 
 | | Crystal structure of human FABP4 in complex with a novel inhibitor | | Descriptor: | 1,2-ETHANEDIOL, 2-[(2-phenylphenyl)amino]benzoic acid, Fatty acid-binding protein, ... | | Authors: | Su, H.X, Zhang, X.L, Li, M.J, Xu, Y.C. | | Deposit date: | 2019-12-17 | | Release date: | 2020-04-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Exploration of Fragment Binding Poses Leading to Efficient Discovery of Highly Potent and Orally Effective Inhibitors of FABP4 for Anti-inflammation.
J.Med.Chem., 63, 2020
|
|
7SYY
 
 | | Hendra virus G protein head domain in complex with cross-neutralizing murine antibody hAH1.3 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Antibody hAH1.3 Heavy Chain, ... | | Authors: | Xu, K, Xu, Y. | | Deposit date: | 2021-11-25 | | Release date: | 2022-06-01 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.74 Å) | | Cite: | Potent monoclonal antibody-mediated neutralization of a divergent Hendra virus variant.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7SYZ
 
 | | Hendra virus G protein head domain in complex with cross-neutralizing murine antibody hAH1.3 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Antibody hAH1.3 Heavy Chain, Antibody hAH1.3 light chain, ... | | Authors: | Xu, K, Xu, Y. | | Deposit date: | 2021-11-25 | | Release date: | 2022-06-08 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.86 Å) | | Cite: | Potent monoclonal antibody-mediated neutralization of a divergent Hendra virus variant.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
6L1K
 
 | | Crystal structure of NADH-dependent butanol dehydrogenase A from Fusobacterium nucleatum | | Descriptor: | NADH-dependent butanol dehydrogenase A, PHOSPHATE ION | | Authors: | Lan, J, Shang, F, Liu, W, Xu, Y, Chen, Y. | | Deposit date: | 2019-09-29 | | Release date: | 2019-10-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Crystal structure of NADH-dependent butanol dehydrogenase A from Fusobacterium nucleatum
To Be Published
|
|
6L19
 
 | | The crystal structure of competence or damage-inducible protein from Enterobacter asburiae | | Descriptor: | CHLORIDE ION, GLYCEROL, PncC family amidohydrolase, ... | | Authors: | Wang, L, Chen, Y, Liu, W, Lan, J, Shang, F, Xu, Y. | | Deposit date: | 2019-09-28 | | Release date: | 2019-10-16 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | The crystal structure of Competence or damage-inducible protein from Enterobacter asburiae
To Be Published
|
|
6LJX
 
 | | Crystal structure of human FABP4 in complex with a novel inhibitor | | Descriptor: | 2-phenylazanylbenzoic acid, Fatty acid-binding protein, adipocyte | | Authors: | Su, H.X, Zhang, X.L, Li, M.J, Xu, Y.C. | | Deposit date: | 2019-12-17 | | Release date: | 2020-04-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Exploration of Fragment Binding Poses Leading to Efficient Discovery of Highly Potent and Orally Effective Inhibitors of FABP4 for Anti-inflammation.
J.Med.Chem., 63, 2020
|
|
6LJU
 
 | | Crystal structure of human FABP4 in complex with a novel inhibitor | | Descriptor: | 1,2-ETHANEDIOL, 2-[[3-chloranyl-4-(methylamino)-2-phenyl-phenyl]amino]benzoic acid, Fatty acid-binding protein, ... | | Authors: | Su, H.X, Zhang, X.L, Li, M.J, Xu, Y.C. | | Deposit date: | 2019-12-17 | | Release date: | 2020-04-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Exploration of Fragment Binding Poses Leading to Efficient Discovery of Highly Potent and Orally Effective Inhibitors of FABP4 for Anti-inflammation.
J.Med.Chem., 63, 2020
|
|
6LJV
 
 | | Crystal structure of human FABP4 in complex with a novel inhibitor | | Descriptor: | 2-[[3-chloranyl-2-(2,3-dihydro-1-benzofuran-5-yl)phenyl]amino]benzoic acid, Fatty acid-binding protein, adipocyte | | Authors: | Su, H.X, Zhang, X.L, Li, M.J, Xu, Y.C. | | Deposit date: | 2019-12-17 | | Release date: | 2020-04-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.401 Å) | | Cite: | Exploration of Fragment Binding Poses Leading to Efficient Discovery of Highly Potent and Orally Effective Inhibitors of FABP4 for Anti-inflammation.
J.Med.Chem., 63, 2020
|
|
6LJW
 
 | | Crystal structure of human FABP4 in complex with a novel inhibitor | | Descriptor: | 1,2-ETHANEDIOL, 2-phenylazanylbenzoic acid, Fatty acid-binding protein, ... | | Authors: | Su, H.X, Zhang, X.L, Li, M.J, Xu, Y.C. | | Deposit date: | 2019-12-17 | | Release date: | 2020-04-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Exploration of Fragment Binding Poses Leading to Efficient Discovery of Highly Potent and Orally Effective Inhibitors of FABP4 for Anti-inflammation.
J.Med.Chem., 63, 2020
|
|
6LJT
 
 | | Crystal structure of human FABP4 in complex with a novel inhibitor | | Descriptor: | 1,2-ETHANEDIOL, 2-[(3-chloranyl-2-phenyl-phenyl)amino]benzoic acid, Fatty acid-binding protein, ... | | Authors: | Su, H.X, Zhang, X.L, Li, M.J, Xu, Y.C. | | Deposit date: | 2019-12-17 | | Release date: | 2020-04-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Exploration of Fragment Binding Poses Leading to Efficient Discovery of Highly Potent and Orally Effective Inhibitors of FABP4 for Anti-inflammation.
J.Med.Chem., 63, 2020
|
|
7BV1
 
 | | Cryo-EM structure of the apo nsp12-nsp7-nsp8 complex | | Descriptor: | Non-structural protein 7, Non-structural protein 8, RNA-directed RNA polymerase, ... | | Authors: | Yin, W, Mao, C, Luan, X, Shen, D, Shen, Q, Su, H, Wang, X, Zhou, F, Zhao, W, Gao, M, Chang, S, Xie, Y.C, Tian, G, Jiang, H.W, Tao, S.C, Shen, J, Jiang, Y, Jiang, H, Xu, Y, Zhang, S, Zhang, Y, Xu, H.E. | | Deposit date: | 2020-04-09 | | Release date: | 2020-04-22 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structural basis for inhibition of the RNA-dependent RNA polymerase from SARS-CoV-2 by remdesivir.
Science, 368, 2020
|
|
7BV2
 
 | | The nsp12-nsp7-nsp8 complex bound to the template-primer RNA and triphosphate form of Remdesivir(RTP) | | Descriptor: | MAGNESIUM ION, Non-structural protein 7, Non-structural protein 8, ... | | Authors: | Yin, W, Mao, C, Luan, X, Shen, D, Shen, Q, Su, H, Wang, X, Zhou, F, Zhao, W, Gao, M, Chang, S, Xie, Y.C, Tian, G, Jiang, H.W, Tao, S.C, Shen, J, Jiang, Y, Jiang, H, Xu, Y, Zhang, S, Zhang, Y, Xu, H.E. | | Deposit date: | 2020-04-09 | | Release date: | 2020-04-22 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Structural basis for inhibition of the RNA-dependent RNA polymerase from SARS-CoV-2 by remdesivir.
Science, 368, 2020
|
|
6LRL
 
 | | Human cGAS catalytic domain bound with compound s2 | | Descriptor: | 3-[[5-(1,2,4-triazol-4-yl)-4H-1,2,4-triazol-3-yl]carbonylamino]benzoic acid, Cyclic GMP-AMP synthase, ZINC ION | | Authors: | Zhao, W.F, Xiong, M.Y, Yuan, X.J, Sun, H.B, Xu, Y.C. | | Deposit date: | 2020-01-16 | | Release date: | 2020-06-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.655 Å) | | Cite: | In Silico Screening-Based Discovery of Novel Inhibitors of Human Cyclic GMP-AMP Synthase: A Cross-Validation Study of Molecular Docking and Experimental Testing.
J.Chem.Inf.Model., 60, 2020
|
|
6LRJ
 
 | | Human cGAS catalytic domain bound with compound 23 | | Descriptor: | 4-[2-(2-methyl-[1,2,4]triazolo[1,5-c]quinazolin-5-yl)hydrazinyl]-4-oxidanylidene-butanoic acid, Cyclic GMP-AMP synthase, ZINC ION | | Authors: | Zhao, W.F, Xiong, M.Y, Yuan, X.J, Sun, H.B, Xu, Y.C. | | Deposit date: | 2020-01-16 | | Release date: | 2020-06-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | In Silico Screening-Based Discovery of Novel Inhibitors of Human Cyclic GMP-AMP Synthase: A Cross-Validation Study of Molecular Docking and Experimental Testing.
J.Chem.Inf.Model., 60, 2020
|
|
