7WPI
 
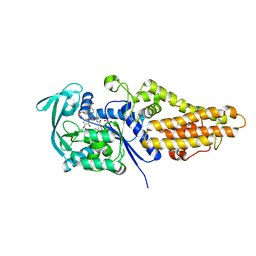 | | Methionyl-tRNA synthetase from Staphylococcus aureus complexed with a phenylbenzimidazole inhibitor | | Descriptor: | 1,2-ETHANEDIOL, 2-(4-bromanyl-3-oxidanyl-phenyl)-1-[(1R)-2-[2-(2-tert-butylphenoxy)ethanoylamino]-1-(3,4-dimethoxyphenyl)ethyl]-N-methyl-benzimidazole-5-carboxamide, ACETATE ION, ... | | Authors: | Yi, J, Cai, Z, Qiu, H, Lu, F, Chen, B, Luo, Z, Gu, Q, Xu, J, Zhou, H. | | Deposit date: | 2022-01-24 | | Release date: | 2022-04-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Fragment screening and structural analyses highlight the ATP-assisted ligand binding for inhibitor discovery against type 1 methionyl-tRNA synthetase.
Nucleic Acids Res., 50, 2022
|
|
7WPM
 
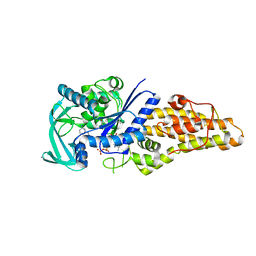 | | Methionyl-tRNA synthetase from Staphylococcus aureus complexed with a fragment and ATP | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5'-TRIPHOSPHATE, Methionine--tRNA ligase, ... | | Authors: | Yi, J, Cai, Z, Qiu, H, Lu, F, Chen, B, Luo, Z, Gu, Q, Xu, J, Zhou, H. | | Deposit date: | 2022-01-24 | | Release date: | 2022-04-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Fragment screening and structural analyses highlight the ATP-assisted ligand binding for inhibitor discovery against type 1 methionyl-tRNA synthetase.
Nucleic Acids Res., 50, 2022
|
|
7WPX
 
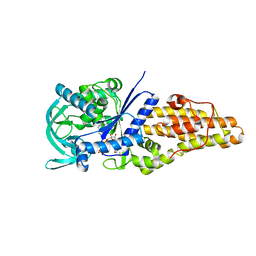 | | Methionyl-tRNA synthetase from Staphylococcus aureus complexed with a fragment and ATP | | Descriptor: | 1,2-ETHANEDIOL, 6-chloranylimidazo[2,1-b][1,3]thiazole, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Yi, J, Cai, Z, Qiu, H, Lu, F, Chen, B, Luo, Z, Gu, Q, Xu, J, Zhou, H. | | Deposit date: | 2022-01-24 | | Release date: | 2022-04-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Fragment screening and structural analyses highlight the ATP-assisted ligand binding for inhibitor discovery against type 1 methionyl-tRNA synthetase.
Nucleic Acids Res., 50, 2022
|
|
7WPN
 
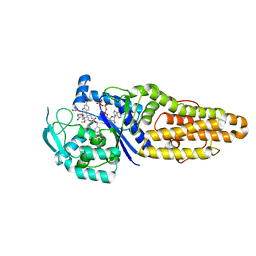 | | Methionyl-tRNA synthetase from Staphylococcus aureus complexed with a phenylbenzimidazole inhibitor and ATP | | Descriptor: | (phenylmethyl) N-[(2R)-2-[2-(4-bromanyl-3-oxidanyl-phenyl)-5-(methylcarbamoyl)benzimidazol-1-yl]-2-(3,4-dimethoxyphenyl)ethyl]carbamate, 1,2-ETHANEDIOL, ACETIC ACID, ... | | Authors: | Yi, J, Cai, Z, Qiu, H, Lu, F, Chen, B, Luo, Z, Gu, Q, Xu, J, Zhou, H. | | Deposit date: | 2022-01-24 | | Release date: | 2022-04-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Fragment screening and structural analyses highlight the ATP-assisted ligand binding for inhibitor discovery against type 1 methionyl-tRNA synthetase.
Nucleic Acids Res., 50, 2022
|
|
7WQ0
 
 | | Methionyl-tRNA synthetase from Staphylococcus aureus complexed with a fragment and ATP | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5'-TRIPHOSPHATE, Methionine--tRNA ligase, ... | | Authors: | Yi, J, Cai, Z, Qiu, H, Lu, F, Chen, B, Luo, Z, Gu, Q, Xu, J, Zhou, H. | | Deposit date: | 2022-01-24 | | Release date: | 2022-04-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Fragment screening and structural analyses highlight the ATP-assisted ligand binding for inhibitor discovery against type 1 methionyl-tRNA synthetase.
Nucleic Acids Res., 50, 2022
|
|
7WPL
 
 | | Methionyl-tRNA synthetase from Staphylococcus aureus complexed with ATP | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5'-TRIPHOSPHATE, Methionine--tRNA ligase | | Authors: | Yi, J, Cai, Z, Qiu, H, Lu, F, Chen, B, Luo, Z, Gu, Q, Xu, J, Zhou, H. | | Deposit date: | 2022-01-24 | | Release date: | 2022-04-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Fragment screening and structural analyses highlight the ATP-assisted ligand binding for inhibitor discovery against type 1 methionyl-tRNA synthetase.
Nucleic Acids Res., 50, 2022
|
|
7W6K
 
 | | Cryo-EM structure of GmALMT12/QUAC1 anion channel | | Descriptor: | GmALMT12/QUAC1 | | Authors: | Qin, L, Tang, L.H, Xu, J.S, Zhang, X.H, Zhu, Y, Sun, F, Su, M, Zhai, Y.J, Chen, Y.H. | | Deposit date: | 2021-12-01 | | Release date: | 2022-03-16 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Cryo-EM structure and electrophysiological characterization of ALMT from Glycine max reveal a previously uncharacterized class of anion channels.
Sci Adv, 8, 2022
|
|
3UUE
 
 | | Crystal structure of mono- and diacylglycerol lipase from Malassezia globosa | | Descriptor: | CHLORIDE ION, GLYCEROL, LIP1, ... | | Authors: | Xu, T, Xu, J, Hou, S, Liu, J. | | Deposit date: | 2011-11-28 | | Release date: | 2012-04-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Crystal structure of a mono- and diacylglycerol lipase from Malassezia globosa reveals a novel lid conformation and insights into the substrate specificity.
J.Struct.Biol., 178, 2012
|
|
5WZT
 
 | | Crystal structure of human secreted phospholipase A2 group IIE with Compound 14 | | Descriptor: | 2-[1-[(3-bromophenyl)methyl]-2-methyl-3-oxamoyl-indol-4-yl]oxyethanoic acid, CALCIUM ION, DIMETHYL SULFOXIDE, ... | | Authors: | Hou, S, Xu, J, Xu, T, Liu, J. | | Deposit date: | 2017-01-18 | | Release date: | 2018-01-24 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis for functional selectivity and ligand recognition revealed by crystal structures of human secreted phospholipase A2 group IIE
Sci Rep, 7, 2017
|
|
5WZO
 
 | | Crystal structure of human secreted phospholipase A2 group IIE, crystallized with calcium | | Descriptor: | 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Hou, S, Xu, J, Xu, T, Liu, J. | | Deposit date: | 2017-01-18 | | Release date: | 2018-01-24 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis for functional selectivity and ligand recognition revealed by crystal structures of human secreted phospholipase A2 group IIE
Sci Rep, 7, 2017
|
|
5WZU
 
 | | Crystal structure of human secreted phospholipase A2 group IIE with Compound 24 | | Descriptor: | 2-[2-methyl-3-oxamoyl-1-[[2-(trifluoromethyl)phenyl]methyl]indol-4-yl]oxyethanoic acid, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Hou, S, Xu, J, Xu, T, Liu, J. | | Deposit date: | 2017-01-18 | | Release date: | 2018-01-24 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis for functional selectivity and ligand recognition revealed by crystal structures of human secreted phospholipase A2 group IIE
Sci Rep, 7, 2017
|
|
5WZW
 
 | | Crystal structure of human secreted phospholipase A2 group IIE with LY311727 | | Descriptor: | (3-{[3-(2-amino-2-oxoethyl)-1-benzyl-2-ethyl-1H-indol-5-yl]oxy}propyl)phosphonic acid, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Hou, S, Xu, J, Xu, T, Liu, J. | | Deposit date: | 2017-01-18 | | Release date: | 2018-01-24 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural basis for functional selectivity and ligand recognition revealed by crystal structures of human secreted phospholipase A2 group IIE
Sci Rep, 7, 2017
|
|
3UUF
 
 | | Crystal structure of mono- and diacylglycerol lipase from Malassezia globosa | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, LIP1, secretory lipase (Family 3), ... | | Authors: | Xu, T, Xu, J, Hou, S, Liu, J. | | Deposit date: | 2011-11-28 | | Release date: | 2012-04-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of a mono- and diacylglycerol lipase from Malassezia globosa reveals a novel lid conformation and insights into the substrate specificity.
J.Struct.Biol., 178, 2012
|
|
5WZM
 
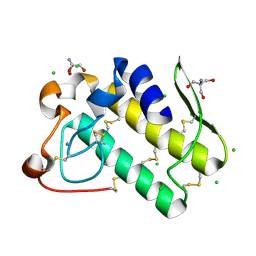 | | Crystal structure of human secreted phospholipase A2 group IIE | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Hou, S, Xu, J, Xu, T, Liu, J. | | Deposit date: | 2017-01-18 | | Release date: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis for functional selectivity and ligand recognition revealed by crystal structures of human secreted phospholipase A2 group IIE
Sci Rep, 7, 2017
|
|
5WZS
 
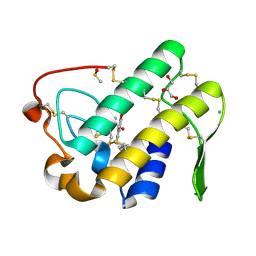 | | Crystal structure of human secreted phospholipase A2 group IIE with Compound 8 | | Descriptor: | 2-[2-methyl-1-(naphthalen-1-ylmethyl)-3-oxamoyl-indol-4-yl]oxyethanoic acid, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Hou, S, Xu, J, Xu, T, Liu, J. | | Deposit date: | 2017-01-18 | | Release date: | 2018-01-24 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis for functional selectivity and ligand recognition revealed by crystal structures of human secreted phospholipase A2 group IIE
Sci Rep, 7, 2017
|
|
5WZV
 
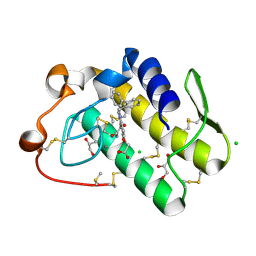 | | Crystal structure of human secreted phospholipase A2 group IIE with Me-indoxam | | Descriptor: | 2-[2-methyl-3-oxamoyl-1-[(2-phenylphenyl)methyl]indol-4-yl]oxyethanoic acid, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Hou, S, Xu, J, Xu, T, Liu, J. | | Deposit date: | 2017-01-18 | | Release date: | 2018-01-24 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis for functional selectivity and ligand recognition revealed by crystal structures of human secreted phospholipase A2 group IIE
Sci Rep, 7, 2017
|
|
4ON3
 
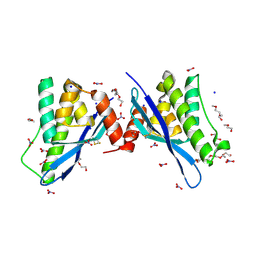 | | Crystal structure of human sorting nexin 10 (SNX10) | | Descriptor: | NITRATE ION, PENTAETHYLENE GLYCOL, SODIUM ION, ... | | Authors: | Xu, T, Xu, J, Wang, Q, Liu, J. | | Deposit date: | 2014-01-28 | | Release date: | 2014-09-24 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of human SNX10 reveals insights into its role in human autosomal recessive osteopetrosis.
Proteins, 82, 2014
|
|
5Y5E
 
 | | Crystal structure of phospholipase A2 with inhibitor | | Descriptor: | 2-[2-methyl-3-oxamoyl-1-[[2-(trifluoromethyl)phenyl]methyl]indol-4-yl]oxyethanoic acid, 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CALCIUM ION, ... | | Authors: | Hou, S, Xu, J, Xu, T, Liu, J. | | Deposit date: | 2017-08-08 | | Release date: | 2018-06-20 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis for functional selectivity and ligand recognition revealed by crystal structures of human secreted phospholipase A2group IIE
Sci Rep, 7, 2017
|
|
8RWJ
 
 | | cryoEM structure of Acs1 filament determined by FilamentID | | Descriptor: | Acetyl-coenzyme A synthetase, [[(2~{R},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methoxy-oxidanyl-phosphoryl] ethanoate | | Authors: | Hugener, J, Xu, J, Wettstein, R, Ioannidi, L, Velikov, D, Wollweber, F, Henggeler, A, Matos, J, Pilhofer, M. | | Deposit date: | 2024-02-05 | | Release date: | 2024-06-26 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | FilamentID reveals the composition and function of metabolic enzyme polymers during gametogenesis.
Cell, 187, 2024
|
|
8RWK
 
 | | cryoEM structure of the central Ald4 filament determined by FilamentID | | Descriptor: | NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Potassium-activated aldehyde dehydrogenase, mitochondrial | | Authors: | Hugener, J, Xu, J, Wettstein, R, Ioannidi, L, Velikov, D, Wollweber, F, Henggeler, A, Matos, J, Pilhofer, M. | | Deposit date: | 2024-02-05 | | Release date: | 2024-06-26 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | FilamentID reveals the composition and function of metabolic enzyme polymers during gametogenesis.
Cell, 187, 2024
|
|
4XX5
 
 | | Structure of PI3K gamma in complex with an inhibitor | | Descriptor: | N-[5-(5-methoxypyridin-3-yl)-4,5,6,7-tetrahydro[1,3]thiazolo[5,4-c]pyridin-2-yl]acetamide, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit gamma isoform | | Authors: | Collier, P.N, Messersmith, D, Le Tiran, A, Bandarage, U.K, Boucher, C, Come, J, Cottrell, K.M, Damagnez, V, Doran, J.D, Griffith, J.P, Khare-Pandit, S, Krueger, E.B, Ledeboer, M.W, Ledford, B, Liao, Y, Mahajan, S, Moody, C.S, Wang, T, Xu, J, Aronov, A.M. | | Deposit date: | 2015-01-29 | | Release date: | 2015-08-05 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.76 Å) | | Cite: | Structure of PI3K gamma in complex with an inhibitor
To Be Published
|
|
4XZ4
 
 | | Structure of PI3K gamma in complex with an inhibitor | | Descriptor: | N-[5-(6-methoxypyrazin-2-yl)-4,5,6,7-tetrahydro[1,3]thiazolo[5,4-c]pyridin-2-yl]acetamide, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit gamma isoform | | Authors: | Collier, P.N, Messersmith, D, Le Tiran, A, Bandarage, U.K, Boucher, C, Come, J, Cottrell, K.M, Damagnez, V, Doran, J.D, Griffith, J.P, Khare-Pandit, S, Krueger, E.B, Ledeboer, M.W, Ledford, B, Liao, Y, Mahajan, S, Moody, C.S, Wang, T, Xu, J, Aronov, A.M. | | Deposit date: | 2015-02-03 | | Release date: | 2016-02-03 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of PI3K gamma in complex with an inhibitor
To Be Published
|
|
8RB4
 
 | |
7V7B
 
 | |
7V7C
 
 | | CryoEM structure of DDB1-VprBP-Vpr-UNG2(94-313) complex | | Descriptor: | DDB1- and CUL4-associated factor 1, DNA damage-binding protein 1, Protein Vpr, ... | | Authors: | Wang, D, Xu, J, Liu, Q, Xiang, Y. | | Deposit date: | 2021-08-21 | | Release date: | 2022-08-31 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structural insights into the HIV-1 Vpr mediated ubiquitination through the Cullin-RING E3 ubiquitin ligase
To Be Published
|
|
