3HNC
 
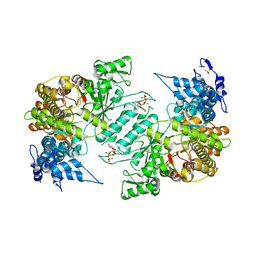 | | Crystal structure of human ribonucleotide reductase 1 bound to the effector TTP | | Descriptor: | MAGNESIUM ION, Ribonucleoside-diphosphate reductase large subunit, SULFATE ION, ... | | Authors: | Fairman, J.W, Wijerathna, S.R, Xu, H, Dealwis, C.G. | | Deposit date: | 2009-05-31 | | Release date: | 2011-02-23 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | Structural basis for allosteric regulation of human ribonucleotide reductase by nucleotide-induced oligomerization.
Nat.Struct.Mol.Biol., 18, 2011
|
|
4ZRD
 
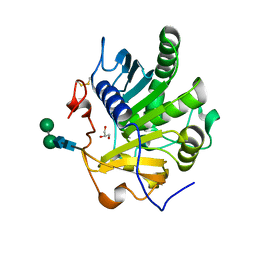 | | Crystal structure of SMG1 F278N mutant | | Descriptor: | GLYCEROL, LIP1, secretory lipase (Family 3), ... | | Authors: | Xu, J, Xu, H, Hou, S, Liu, J. | | Deposit date: | 2015-05-12 | | Release date: | 2015-09-23 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of product-bound SMG1 lipase: active site gating implications.
Febs J., 282, 2015
|
|
4ZRE
 
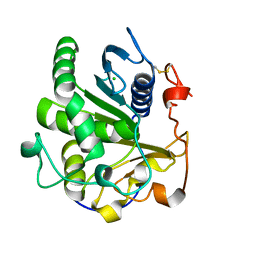 | | Crystal structure of SMG1 F278D mutant | | Descriptor: | CHLORIDE ION, LIP1, secretory lipase (Family 3), ... | | Authors: | Xu, J, Xu, H, Hou, S, Liu, J. | | Deposit date: | 2015-05-12 | | Release date: | 2015-09-23 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of product-bound SMG1 lipase: active site gating implications.
Febs J., 282, 2015
|
|
1YP0
 
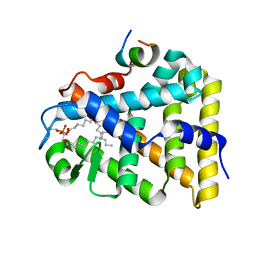 | | Structure of the steroidogenic factor-1 ligand binding domain bound to phospholipid and a SHP peptide motif | | Descriptor: | DI-PALMITOYL-3-SN-PHOSPHATIDYLETHANOLAMINE, Nuclear receptor subfamily 0, group B, ... | | Authors: | Li, Y, Choi, M, Cavey, G, Daugherty, J, Suino, K, Kovach, A, Bingham, N, Kliewer, S, Xu, H. | | Deposit date: | 2005-01-28 | | Release date: | 2005-04-19 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystallographic identification and functional characterization of phospholipids as ligands for the orphan nuclear receptor steroidogenic factor-1.
Mol.Cell, 17, 2005
|
|
6J4G
 
 | | Crystal structure of the AtWRKY33 domain | | Descriptor: | DNA (5'-D(*AP*GP*CP*CP*TP*TP*TP*GP*AP*CP*CP*AP*GP*CP*G)-3'), DNA (5'-D(*TP*CP*GP*CP*TP*GP*GP*TP*CP*AP*AP*AP*GP*GP*C)-3'), Probable WRKY transcription factor 33, ... | | Authors: | Xu, Y.P, Xu, H, Wang, B, Su, X.D. | | Deposit date: | 2019-01-08 | | Release date: | 2020-01-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of the N-terminal DNA binding domain of AtWRKY33
To Be Published
|
|
3GED
 
 | | Fingerprint and Structural Analysis of a Apo SCOR enzyme from Clostridium thermocellum | | Descriptor: | GLYCEROL, SODIUM ION, Short-chain dehydrogenase/reductase SDR, ... | | Authors: | Huether, R, Liu, Z.J, Xu, H, Wang, B.C, Pletnev, V, Mao, Q, Umland, T, Duax, W. | | Deposit date: | 2009-02-25 | | Release date: | 2009-03-17 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.698 Å) | | Cite: | Sequence fingerprint and structural analysis of the SCOR enzyme A3DFK9 from Clostridium thermocellum.
Proteins, 78, 2010
|
|
3GEG
 
 | | Fingerprint and Structural Analysis of a SCOR enzyme with its bound cofactor from Clostridium thermocellum | | Descriptor: | GLYCEROL, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, SODIUM ION, ... | | Authors: | Huether, R, Liu, Z.J, Xu, H, Wang, B.C, Pletnev, V, Mao, Q, Umland, T, Duax, W. | | Deposit date: | 2009-02-25 | | Release date: | 2009-03-17 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.102 Å) | | Cite: | Sequence fingerprint and structural analysis of the SCOR enzyme A3DFK9 from Clostridium thermocellum.
Proteins, 78, 2010
|
|
3C3D
 
 | | Crystal structure of 2-phospho-(S)-lactate transferase from Methanosarcina mazei in complex with Fo and phosphate. Northeast Structural Genomics Consortium target MaR46 | | Descriptor: | 1-deoxy-1-(8-hydroxy-2,4-dioxo-3,4-dihydropyrimido[4,5-b]quinolin-10(2H)-yl)-D-ribitol, 2-phospho-L-lactate transferase, PHOSPHATE ION | | Authors: | Forouhar, F, Abashidze, M, Xu, H, Grochowski, L.L, Seetharaman, J, Hussain, M, Kuzin, A.P, Chen, Y, Zhou, W, Xiao, R, Acton, T.B, Montelione, G.T, Galinier, A, White, R.H, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2008-01-28 | | Release date: | 2008-02-19 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Molecular insights into the biosynthesis of the f420 coenzyme.
J.Biol.Chem., 283, 2008
|
|
6J4E
 
 | | Crystal structure of the AtWRKY1 domain | | Descriptor: | DNA (5'-D(*AP*GP*CP*CP*TP*TP*TP*GP*AP*CP*CP*AP*GP*CP*G)-3'), DNA (5'-D(*TP*CP*GP*CP*TP*GP*GP*TP*CP*AP*AP*AP*GP*GP*C)-3'), WRKY transcription factor 1, ... | | Authors: | Xu, Y.P, Xu, H, Wang, B, Su, X.D. | | Deposit date: | 2019-01-08 | | Release date: | 2020-01-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.126 Å) | | Cite: | Crystal structure of the AtWRKY1 domain
To Be Published
|
|
3C3E
 
 | | Crystal structure of 2-phospho-(S)-lactate transferase from Methanosarcina mazei in complex with Fo and GDP. Northeast Structural Genomics Consortium target MaR46 | | Descriptor: | 1-deoxy-1-(8-hydroxy-2,4-dioxo-3,4-dihydropyrimido[4,5-b]quinolin-10(2H)-yl)-D-ribitol, 2-phospho-L-lactate transferase, GUANOSINE-5'-DIPHOSPHATE | | Authors: | Forouhar, F, Abashidze, M, Xu, H, Grochowski, L.L, Seetharaman, J, Hussain, M, Kuzin, A.P, Chen, Y, Zhou, W, Xiao, R, Acton, T.B, Montelione, G.T, Galinier, A, White, R.H, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2008-01-28 | | Release date: | 2008-02-19 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Molecular insights into the biosynthesis of the f420 coenzyme.
J.Biol.Chem., 283, 2008
|
|
3BZU
 
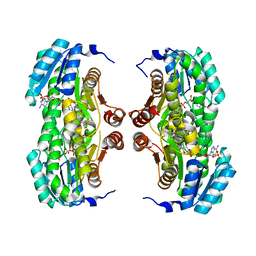 | | Crystal structure of human 11-beta-hydroxysteroid dehydrogenase(HSD1) in complex with NADP and thiazolone inhibitor | | Descriptor: | (5S)-2-{[(1S)-1-(2-fluorophenyl)ethyl]amino}-5-methyl-5-(trifluoromethyl)-1,3-thiazol-4(5H)-one, Corticosteroid 11-beta-dehydrogenase isozyme 1, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Min, X, Sudom, A, Xu, H, Wang, Z, Walker, N.P. | | Deposit date: | 2008-01-18 | | Release date: | 2008-06-10 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural characterization and pharmacodynamic effects of an orally active 11beta-hydroxysteroid dehydrogenase type 1 inhibitor.
Chem.Biol.Drug Des., 71, 2008
|
|
6J4F
 
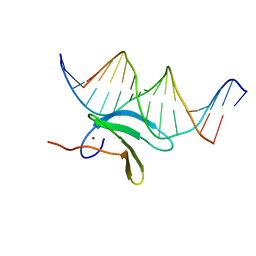 | | Crystal structure of the AtWRKY2 domain | | Descriptor: | DNA (5'-D(*AP*GP*CP*CP*TP*TP*TP*GP*AP*CP*CP*AP*GP*CP*G)-3'), DNA (5'-D(*TP*CP*GP*CP*TP*GP*GP*TP*CP*AP*AP*AP*GP*GP*C)-3'), Probable WRKY transcription factor 2, ... | | Authors: | Xu, Y.P, Xu, H, Wang, B, Su, X.D. | | Deposit date: | 2019-01-08 | | Release date: | 2020-01-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of the AtWRKY2 domain
To Be Published
|
|
2IDG
 
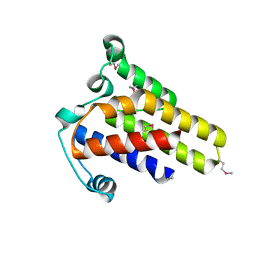 | | Crystal Structure of hypothetical protein AF0160 from Archaeoglobus fulgidus | | Descriptor: | Hypothetical protein AF0160 | | Authors: | Zhao, M, Zhang, M, Chang, J, Chen, L, Xu, H, Li, Y, Liu, Z.J, Rose, J.P, Wang, B.C, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2006-09-15 | | Release date: | 2006-11-14 | | Last modified: | 2017-09-13 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | Crystal structure of Hypothetical Protein AF0160 from Archaeoglobus fulgidus at 2.69 Angstrom resolution
To be Published
|
|
1SGL
 
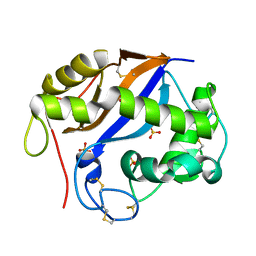 | | The three-dimensional structure and X-ray sequence reveal that trichomaglin is a novel S-like ribonuclease | | Descriptor: | SULFATE ION, trichomaglin | | Authors: | Gan, J.-H, Yu, L, Wu, J, Xu, H, Choudhary, J.S, Blackstock, W.P, Liu, W.-Y, Xia, Z.-X. | | Deposit date: | 2004-02-24 | | Release date: | 2004-06-22 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The three-dimensional structure and X-ray sequence reveal that trichomaglin is a novel S-like ribonuclease.
Structure, 12, 2004
|
|
2IEL
 
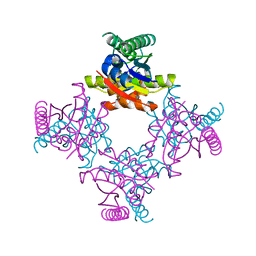 | | CRYSTAL STRUCTURE OF TT0030 from Thermus Thermophilus | | Descriptor: | Hypothetical Protein TT0030 | | Authors: | Zhu, J, Huang, J, Stepanyuk, G, Chen, L, Chang, J, Zhao, M, Xu, H, Liu, Z.J, Rose, J.P, Wang, B.C, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2006-09-19 | | Release date: | 2006-11-14 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | CRYSTAL STRUCTURE OF TT0030 from Thermus Thermophilus AT 1.6 ANGSTROMS RESOLUTION
To be Published
|
|
2O4X
 
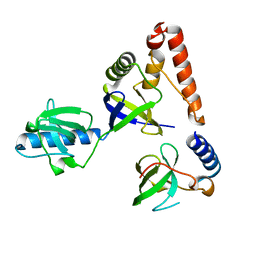 | | Crystal structure of human P100 tudor domain | | Descriptor: | Staphylococcal nuclease domain-containing protein 1 | | Authors: | Shaw, N, Zhao, M, Cheng, C, Xu, H, Yang, J, Silvennoinen, O, Rao, Z, Wang, B.C, Liu, Z.J. | | Deposit date: | 2006-12-05 | | Release date: | 2007-02-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of human P100 tudor domain
To be Published
|
|
2HQ4
 
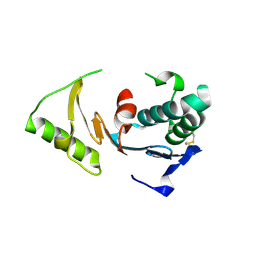 | | Crystal Structure of ORF 1580 a hypothetical protein from Pyrococcus horikoshii | | Descriptor: | Hypothetical protein PH1570 | | Authors: | Li, Y, Marshall, M, Chang, J, Zhao, M, Zhang, M, Xu, H, Liu, Z.J, Rose, J.P, Wang, B.C, Southeast Collaboratory for Structural Genomics, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2006-07-18 | | Release date: | 2006-09-19 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Crystal Structure of ORF 1580 a hypothetical protein from Pyrococcus horikoshii
To be Published
|
|
2HQ1
 
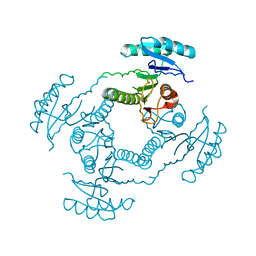 | | Crystal Structure of ORF 1438 a putative Glucose/ribitol dehydrogenase from Clostridium thermocellum | | Descriptor: | Glucose/ribitol dehydrogenase | | Authors: | Southeast Collaboratory for Structural Genomics (SECSG), Li, Y, Shaw, N, Xu, H, Cheng, C, Chen, L, Liu, Z.J, Rose, J.P, Wang, B.C. | | Deposit date: | 2006-07-18 | | Release date: | 2006-09-12 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure of ORF 1438 a putative Glucose/ ribitol dehydrogenase from Clostridium thermocellum
To be Published
|
|
2PL5
 
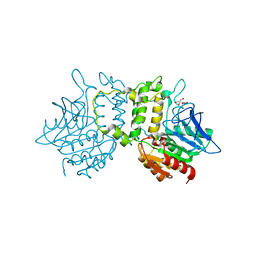 | | Crystal Structure of Homoserine O-acetyltransferase from Leptospira interrogans | | Descriptor: | GLYCEROL, Homoserine O-acetyltransferase | | Authors: | Liu, L, Wang, M, Wang, Y, Wei, Z, Xu, H, Gong, W. | | Deposit date: | 2007-04-19 | | Release date: | 2007-11-20 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of homoserine O-acetyltransferase from Leptospira interrogans
Biochem.Biophys.Res.Commun., 363, 2007
|
|
2HQE
 
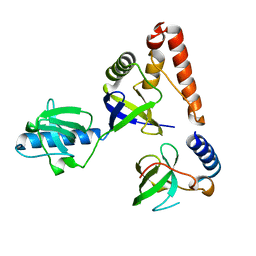 | | Crystal structure of human P100 Tudor domain: Large fragment | | Descriptor: | P100 Co-activator tudor domain | | Authors: | Shah, N, Zhao, M, Cheng, C, Xu, H, Yang, J, Silvennoinen, O, Liu, Z.J, Wang, B.C, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2006-07-18 | | Release date: | 2007-07-03 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of a large fragment of the Human P100 Tudor Domain
To be Published
|
|
2HQX
 
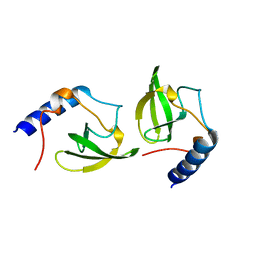 | | Crystal structure of human P100 tudor domain conserved region | | Descriptor: | P100 CO-ACTIVATOR TUDOR DOMAIN | | Authors: | Zhao, M, Liu, Z.J, Xu, H, Yang, J, Silvennoinen, O, Wang, B.C, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2006-07-19 | | Release date: | 2006-10-10 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Crystal Structure of Human P100 Tudor Domain Conserved Region
To be Published
|
|
2QRK
 
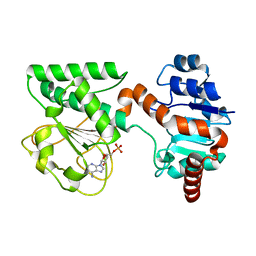 | | Crystal Structure of AMP-bound Saccharopine Dehydrogenase (L-Lys Forming) from Saccharomyces cerevisiae | | Descriptor: | ADENOSINE MONOPHOSPHATE, Saccharopine dehydrogenase [NAD+, L-lysine-forming | | Authors: | Andi, B, Xu, H, Cook, P.F, West, A.H. | | Deposit date: | 2007-07-28 | | Release date: | 2007-10-30 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal Structures of Ligand-Bound Saccharopine Dehydrogenase from Saccharomyces cerevisiae
Biochemistry, 46, 2007
|
|
2QRJ
 
 | | Crystal Structure of Sulfate-bound Saccharopine Dehydrogenase (L-Lys Forming) from Saccharomyces cerevisiae | | Descriptor: | CHLORIDE ION, SULFATE ION, Saccharopine dehydrogenase, ... | | Authors: | Andi, B, Xu, H, Cook, P.F, West, A.H. | | Deposit date: | 2007-07-28 | | Release date: | 2007-10-30 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal Structures of Ligand-Bound Saccharopine Dehydrogenase from Saccharomyces cerevisiae
Biochemistry, 46, 2007
|
|
2QK8
 
 | | Crystal structure of the anthrax drug target, Bacillus anthracis dihydrofolate reductase | | Descriptor: | Dihydrofolate reductase, METHOTREXATE | | Authors: | Bennett, B.C, Xu, H, Simmerman, R.F, Lee, R.E, Dealwis, C.G. | | Deposit date: | 2007-07-10 | | Release date: | 2007-08-28 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of the anthrax drug target, Bacillus anthracis dihydrofolate reductase.
J.Med.Chem., 50, 2007
|
|
2QRL
 
 | | Crystal Structure of Oxalylglycine-bound Saccharopine Dehydrogenase (L-Lys Forming) from Saccharomyces cerevisiae | | Descriptor: | N-OXALYLGLYCINE, Saccharopine dehydrogenase, NAD+, ... | | Authors: | Andi, B, Xu, H, Cook, P.F, West, A.H. | | Deposit date: | 2007-07-28 | | Release date: | 2007-10-30 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal Structures of Ligand-Bound Saccharopine Dehydrogenase from Saccharomyces cerevisiae
Biochemistry, 46, 2007
|
|
