7DCM
 
 | | Crystal structure of CITX | | Descriptor: | Probable apo-citrate lyase phosphoribosyl-dephospho-CoA transferase, ZINC ION | | Authors: | Xu, H, Wang, B, Su, X.D. | | Deposit date: | 2020-10-26 | | Release date: | 2021-11-03 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.495 Å) | | Cite: | Co-evolution-based prediction of metal-binding sites in proteomes by machine learning.
Nat.Chem.Biol., 19, 2023
|
|
7EK0
 
 | | Complex Structure of antibody BD-503 and RBD-N501Y of COVID-19 | | Descriptor: | Heavy Chain of BD-503, Light Chain of BD-503, Spike protein S1 | | Authors: | Xu, H, Wang, B, Zhao, T.N, Su, X.D. | | Deposit date: | 2021-04-03 | | Release date: | 2022-04-13 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure-based analyses of neutralization antibodies interacting with naturally occurring SARS-CoV-2 RBD variants.
Cell Res., 31, 2021
|
|
7EJY
 
 | | Complex Structure of antibody BD-503 and RBD of COVID-19 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy Chain of BD-503, Light Chain of BD-503, ... | | Authors: | Xu, H, Wang, B, Zhao, T.N, Su, X.D. | | Deposit date: | 2021-04-03 | | Release date: | 2022-04-13 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.04 Å) | | Cite: | Structure-based analyses of neutralization antibodies interacting with naturally occurring SARS-CoV-2 RBD variants.
Cell Res., 31, 2021
|
|
7EJZ
 
 | | Complex Structure of antibody BD-503 and RBD-S477N of COVID-19 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy Chain of BD-503, Light Chain of BD-503, ... | | Authors: | Xu, H, Wang, B, Zhao, T.N, Su, X.D. | | Deposit date: | 2021-04-03 | | Release date: | 2022-04-13 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.63 Å) | | Cite: | Structure-based analyses of neutralization antibodies interacting with naturally occurring SARS-CoV-2 RBD variants.
Cell Res., 31, 2021
|
|
7F6Z
 
 | | Complex Structure of antibody BD-503 and RBD-501Y.V2 of COVID-19 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy Chain of BD-503, Light Chain of BD-503, ... | | Authors: | Xu, H, Wang, B, Zhao, T.N, Su, X.D. | | Deposit date: | 2021-06-26 | | Release date: | 2022-06-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure-based analyses of neutralization antibodies interacting with naturally occurring SARS-CoV-2 RBD variants.
Cell Res., 31, 2021
|
|
7F6Y
 
 | | Complex Structure of antibody BD-503 and RBD-E484K of COVID-19 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy Chain of BD-503, Light Chain of BD-503, ... | | Authors: | Xu, H, Wang, B, Zhao, T.N, Su, X.D. | | Deposit date: | 2021-06-26 | | Release date: | 2022-06-29 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure-based analyses of neutralization antibodies interacting with naturally occurring SARS-CoV-2 RBD variants.
Cell Res., 31, 2021
|
|
5JA5
 
 | | Crystal structure of the rice Topless related protein 2 (TPR2) N-terminal topless domain (1-209) L111A and L130A mutant in complex with rice D53 repressor EAR peptide motif | | Descriptor: | Protein TPR1, The rice D53 peptide (a.a. 794-808), ZINC ION | | Authors: | Ke, J, Ma, H, Gu, X, Brunzelle, J.S, Xu, H.E, Melcher, K. | | Deposit date: | 2016-04-12 | | Release date: | 2017-07-05 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A D53 repression motif induces oligomerization of TOPLESS corepressors and promotes assembly of a corepressor-nucleosome complex.
Sci Adv, 3, 2017
|
|
3KAZ
 
 | | Crystal structure of abscisic acid receptor PYL2 | | Descriptor: | 1,3-BUTANEDIOL, Putative uncharacterized protein At2g26040 | | Authors: | Zhou, X.E, Melcher, K, Ng, L.-M, Soon, F.-F, Xu, Y, Suino-Powell, K.M, Kovach, A, Li, J, Xu, H.E. | | Deposit date: | 2009-10-19 | | Release date: | 2009-12-08 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Agate-latch-lock mechanism for hormone signalling by abscisic acid receptors
Nature, 462, 2009
|
|
3KB3
 
 | | Crystal structure of abscisic acid-bound PYL2 in complex with HAB1 | | Descriptor: | (2Z,4E)-5-[(1S)-1-hydroxy-2,6,6-trimethyl-4-oxocyclohex-2-en-1-yl]-3-methylpenta-2,4-dienoic acid, MAGNESIUM ION, Protein phosphatase 2C 16, ... | | Authors: | Zhou, X.E, Melcher, K, Soon, F.-F, Ng, L.-M, Xu, Y, Suino-Powell, K.M, Kovach, A, Li, J, Xu, H.E. | | Deposit date: | 2009-10-20 | | Release date: | 2009-12-08 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Agate-latch-lock mechanism for hormone signalling by abscisic acid receptors
Nature, 462, 2009
|
|
3KAY
 
 | | Crystal structure of abscisic acid receptor PYL1 | | Descriptor: | Putative uncharacterized protein At5g46790 | | Authors: | Zhou, X.E, Melcher, K, Ng, L.-M, Soon, F.-F, Xu, Y, Suino-Powell, K.M, Kovach, A, Li, J, Xu, H.E. | | Deposit date: | 2009-10-19 | | Release date: | 2009-12-08 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Agate-latch-lock mechanism for hormone signalling by abscisic acid receptors
Nature, 462, 2009
|
|
3KB0
 
 | | Crystal structure of abscisic acid-bound PYL2 | | Descriptor: | (2Z,4E)-5-[(1S)-1-hydroxy-2,6,6-trimethyl-4-oxocyclohex-2-en-1-yl]-3-methylpenta-2,4-dienoic acid, Putative uncharacterized protein At2g26040 | | Authors: | Zhou, X.E, Melcher, K, Ng, L.-M, Soon, F.-F, Xu, Y, Suino-Powell, K.M, Kovach, A, Li, J, Xu, H.E. | | Deposit date: | 2009-10-19 | | Release date: | 2009-12-08 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Agate-latch-lock mechanism for hormone signalling by abscisic acid receptors
Nature, 462, 2009
|
|
4MY2
 
 | | Crystal Structure of Norrin in fusion with Maltose Binding Protein | | Descriptor: | Maltose-binding periplasmic protein, Norrin fusion protein, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Ke, J, Jurecky, C, Chen, C, Gu, X, Parker, N, Williams, B.O, Melcher, K, Xu, H.E. | | Deposit date: | 2013-09-27 | | Release date: | 2013-11-13 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure and function of Norrin in assembly and activation of a Frizzled 4-Lrp5/6 complex.
Genes Dev., 27, 2013
|
|
6UH3
 
 | | Crystal structure of bacterial heliorhodopsin 48C12 | | Descriptor: | DI(HYDROXYETHYL)ETHER, Heliorhodopsin, PALMITIC ACID, ... | | Authors: | Lu, Y, Zhou, X.E, Gao, X, Xia, R, Xu, Z, Wang, N, Leng, Y, Melcher, K, Xu, H.E, He, Y. | | Deposit date: | 2019-09-26 | | Release date: | 2019-12-04 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of heliorhodopsin 48C12.
Cell Res., 30, 2020
|
|
4LOG
 
 | | The crystal structure of the orphan nuclear receptor PNR ligand binding domain fused with MBP | | Descriptor: | Maltose ABC transporter periplasmic protein and NR2E3 protein chimeric construct | | Authors: | Tan, M.E, Zhou, X.E, Soon, F.-F, Li, X, Li, J, Yong, E.-L, Melcher, K, Xu, H.E. | | Deposit date: | 2013-07-12 | | Release date: | 2013-10-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The Crystal Structure of the Orphan Nuclear Receptor NR2E3/PNR Ligand Binding Domain Reveals a Dimeric Auto-Repressed Conformation.
Plos One, 8, 2013
|
|
5IZQ
 
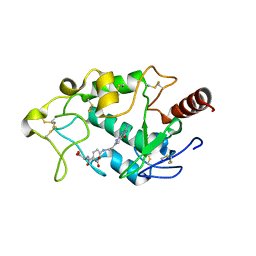 | | Crystal structure of human folate receptor alpha in complex with novel antifolate AGF183 | | Descriptor: | Folate receptor alpha, N-(4-{[2-(2-amino-4-oxo-4,7-dihydro-3H-pyrrolo[2,3-d]pyrimidin-6-yl)ethyl]amino}benzene-1-carbonyl)-L-glutamic acid | | Authors: | Ke, J, Gu, X, Brunzelle, J.S, Xu, H.E, Melcher, K. | | Deposit date: | 2016-03-25 | | Release date: | 2016-08-10 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Tumor Targeting with Novel 6-Substituted Pyrrolo [2,3-d] Pyrimidine Antifolates with Heteroatom Bridge Substitutions via Cellular Uptake by Folate Receptor alpha and the Proton-Coupled Folate Transporter and Inhibition of de Novo Purine Nucleotide Biosynthesis.
J.Med.Chem., 59, 2016
|
|
3C3D
 
 | | Crystal structure of 2-phospho-(S)-lactate transferase from Methanosarcina mazei in complex with Fo and phosphate. Northeast Structural Genomics Consortium target MaR46 | | Descriptor: | 1-deoxy-1-(8-hydroxy-2,4-dioxo-3,4-dihydropyrimido[4,5-b]quinolin-10(2H)-yl)-D-ribitol, 2-phospho-L-lactate transferase, PHOSPHATE ION | | Authors: | Forouhar, F, Abashidze, M, Xu, H, Grochowski, L.L, Seetharaman, J, Hussain, M, Kuzin, A.P, Chen, Y, Zhou, W, Xiao, R, Acton, T.B, Montelione, G.T, Galinier, A, White, R.H, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2008-01-28 | | Release date: | 2008-02-19 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Molecular insights into the biosynthesis of the f420 coenzyme.
J.Biol.Chem., 283, 2008
|
|
7XTB
 
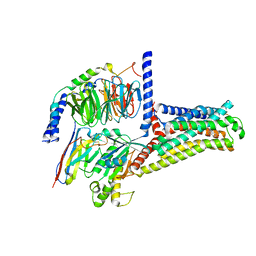 | | Serotonin 6 (5-HT6) receptor-Gs-Nb35 complex | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(s) subunit alpha isoforms short, ... | | Authors: | Huang, S, Xu, P, Shen, D.D, Simon, I.A, Mao, C, Tan, Y, Zhang, H, Harpsoe, K, Li, H, Zhang, Y, You, C, Yu, X, Jiang, Y, Zhang, Y, Gloriam, D.E, Xu, H.E. | | Deposit date: | 2022-05-16 | | Release date: | 2022-07-27 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | GPCRs steer G i and G s selectivity via TM5-TM6 switches as revealed by structures of serotonin receptors.
Mol.Cell, 82, 2022
|
|
7XTA
 
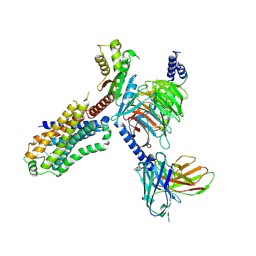 | | Serotonin 4 (5-HT4) receptor-Gi-scFv16 complex | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(i) subunit alpha-1, ... | | Authors: | Huang, S, Xu, P, Shen, D.D, Simon, I.A, Mao, C, Tan, Y, Zhang, H, Harpsoe, K, Li, H, Zhang, Y, You, C, Yu, X, Jiang, Y, Zhang, Y, Gloriam, D.E, Xu, H.E. | | Deposit date: | 2022-05-16 | | Release date: | 2022-07-27 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | GPCRs steer G i and G s selectivity via TM5-TM6 switches as revealed by structures of serotonin receptors.
Mol.Cell, 82, 2022
|
|
7XTC
 
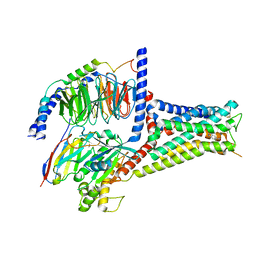 | | Serotonin 7 (5-HT7) receptor-Gs-Nb35 complex | | Descriptor: | 3-(2-azanylethyl)-1H-indole-5-carboxamide, CHOLESTEROL, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Huang, S, Xu, P, Shen, D.D, Simon, I.A, Mao, C, Tan, Y, Zhang, H, Harpsoe, K, Li, H, Zhang, Y, You, C, Yu, X, Jiang, Y, Zhang, Y, Gloriam, D.E, Xu, H.E. | | Deposit date: | 2022-05-16 | | Release date: | 2022-07-27 | | Last modified: | 2022-08-03 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | GPCRs steer G i and G s selectivity via TM5-TM6 switches as revealed by structures of serotonin receptors.
Mol.Cell, 82, 2022
|
|
7XT9
 
 | | Serotonin 4 (5-HT4) receptor-Gs complex | | Descriptor: | CHOLESTEROL, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Huang, S, Xu, P, Shen, D.D, Simon, I.A, Mao, C, Tan, Y, Zhang, H, Harpsoe, K, Li, H, Zhang, Y, You, C, Yu, X, Jiang, Y, Zhang, Y, Gloriam, D.E, Xu, H.E. | | Deposit date: | 2022-05-16 | | Release date: | 2022-07-27 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | GPCRs steer G i and G s selectivity via TM5-TM6 switches as revealed by structures of serotonin receptors.
Mol.Cell, 82, 2022
|
|
7XT8
 
 | | Serotonin 4 (5-HT4) receptor-Gs-Nb35 complex | | Descriptor: | CHOLESTEROL, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Huang, S, Xu, P, Shen, D.D, Simon, I.A, Mao, C, Tan, Y, Zhang, H, Harpsoe, K, Li, H, Zhang, Y, You, C, Yu, X, Jiang, Y, Zhang, Y, Gloriam, D.E, Xu, H.E. | | Deposit date: | 2022-05-16 | | Release date: | 2022-07-27 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | GPCRs steer G i and G s selectivity via TM5-TM6 switches as revealed by structures of serotonin receptors.
Mol.Cell, 82, 2022
|
|
3C0F
 
 | | Crystal Structure of a novel non-Pfam protein AF1514 from Archeoglobus fulgidus DSM 4304 solved by S-SAD using a Cr X-ray source | | Descriptor: | Uncharacterized protein AF_1514 | | Authors: | Li, Y, Bahti, P, Shaw, N, Song, G, Yin, J, Zhu, J.-Y, Zhang, H, Xu, H, Wang, B.-C, Liu, Z.-J. | | Deposit date: | 2008-01-20 | | Release date: | 2008-02-05 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of a novel non-Pfam protein AF1514 from Archeoglobus fulgidus DSM 4304 solved by S-SAD using a Cr X-ray source.
Proteins, 71, 2008
|
|
7XZ5
 
 | | GPR119-Gs-LPC complex | | Descriptor: | (4R,7R,18E)-4,7-dihydroxy-N,N,N-trimethyl-10-oxo-3,5,9-trioxa-4-phosphaheptacos-18-en-1-aminium 4-oxide, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Xu, P, Huang, S, Jiang, Y, Xu, H.E. | | Deposit date: | 2022-06-02 | | Release date: | 2022-08-24 | | Last modified: | 2022-10-05 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural identification of lysophosphatidylcholines as activating ligands for orphan receptor GPR119.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7XZ6
 
 | | GPR119-Gs-APD668 complex | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(s) subunit alpha isoforms short, ... | | Authors: | Xu, P, Huang, S, Jiang, Y, Xu, H.E. | | Deposit date: | 2022-06-02 | | Release date: | 2022-08-24 | | Last modified: | 2022-10-05 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structural identification of lysophosphatidylcholines as activating ligands for orphan receptor GPR119.
Nat.Struct.Mol.Biol., 29, 2022
|
|
8Y53
 
 | | Cryo-EM structure of the MK-5046-bound BRS3-Gq complex | | Descriptor: | (2~{S})-1,1,1-tris(fluoranyl)-2-(4-pyrazol-1-ylphenyl)-3-[5-[[1-(trifluoromethyl)cyclopropyl]methyl]-1~{H}-imidazol-2-yl]propan-2-ol, Bombesin receptor subtype-3, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Li, C, Xu, Y, Yin, W, Xu, H.E. | | Deposit date: | 2024-01-31 | | Release date: | 2024-09-11 | | Method: | ELECTRON MICROSCOPY (2.93 Å) | | Cite: | Structural insights into ligand recognition, selectivity, and activation of bombesin receptor subtype-3.
Cell Rep, 43, 2024
|
|
