6J8O
 
 | | Structure of a hypothetical protease | | Descriptor: | 8-mer peptide, Small vasohibin-binding protein, Tubulinyl-Tyr carboxypeptidase 1 | | Authors: | Liao, S, Gao, J, Xu, C. | | Deposit date: | 2019-01-20 | | Release date: | 2020-01-22 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.855 Å) | | Cite: | Structure of a hypothetical protease
To Be Published
|
|
6J91
 
 | | Structure of a hypothetical protease | | Descriptor: | Small vasohibin-binding protein, Tubulinyl-Tyr carboxypeptidase 1 | | Authors: | Liao, S, Gao, J, Xu, C. | | Deposit date: | 2019-01-21 | | Release date: | 2019-06-19 | | Last modified: | 2019-07-17 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Molecular basis of vasohibins-mediated detyrosination and its impact on spindle function and mitosis.
Cell Res., 29, 2019
|
|
3R93
 
 | | Crystal structure of the chromo domain of M-phase phosphoprotein 8 bound to H3K9Me3 peptide | | Descriptor: | H3K9Me3 peptide, M-phase phosphoprotein 8, UNKNOWN ATOM OR ION | | Authors: | Li, J, Li, Z, Ruan, J, Xu, C, Tong, Y, Pan, P.W, Tempel, W, Crombet, L, Min, J, Zang, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2011-03-24 | | Release date: | 2011-04-06 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.057 Å) | | Cite: | Structural basis for specific binding of human MPP8 chromodomain to histone H3 methylated at lysine 9.
Plos One, 6, 2011
|
|
6IDE
 
 | | Crystal structure of the Vibrio cholera VqmA-Ligand-DNA complex provides molecular mechanisms for drug design | | Descriptor: | 3,5-dimethylpyrazin-2-ol, DNA (5'-D(*AP*GP*GP*GP*GP*GP*GP*AP*AP*AP*TP*CP*CP*CP*CP*CP*CP*T)-3'), DNA (5'-D(*AP*GP*GP*GP*GP*GP*GP*AP*TP*TP*TP*CP*CP*CP*CP*CP*CP*T)-3'), ... | | Authors: | Wu, H, Li, M.J, Guo, H.J, Zhou, H, Li, B, Xu, Q, Xu, C.Y, Yu, F, He, J.H. | | Deposit date: | 2018-09-09 | | Release date: | 2019-01-16 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Crystal structure of theVibrio choleraeVqmA-ligand-DNA complex provides insight into ligand-binding mechanisms relevant for drug design.
J. Biol. Chem., 294, 2019
|
|
6K0X
 
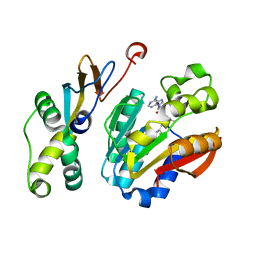 | |
6A9W
 
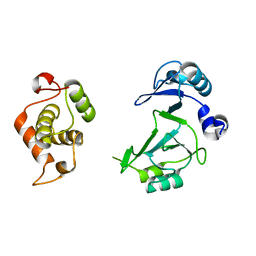 | | Structure of the bifunctional DNA primase-polymerase from phage NrS-1 | | Descriptor: | Primase | | Authors: | Guo, H.J, Li, M.J, Wang, T.L, Wu, H, Zhou, H, Xu, C.Y, Liu, X.P, Yu, F, He, J.H. | | Deposit date: | 2018-07-16 | | Release date: | 2019-03-13 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure and biochemical studies of the bifunctional DNA primase-polymerase from phage NrS-1.
Biochem. Biophys. Res. Commun., 510, 2019
|
|
5X4H
 
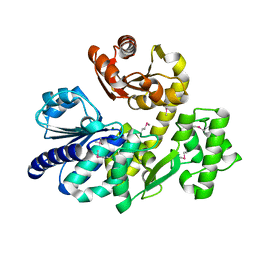 | | The crystal structure of Pyrococcus furiosus RecJ (wild-type) | | Descriptor: | MAGNESIUM ION, Uncharacterized protein | | Authors: | Li, M.J, Yi, G.S, Yu, F, Zhou, H, Chen, J.N, Xu, C.Y, Wang, F.P, Xiao, X, He, J.H, Liu, X.P. | | Deposit date: | 2017-02-13 | | Release date: | 2018-02-14 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.801 Å) | | Cite: | The crystal structure of Pyrococcus furiosus RecJ implicates it as an ancestor of eukaryotic Cdc45.
Nucleic Acids Res., 45, 2017
|
|
5C0M
 
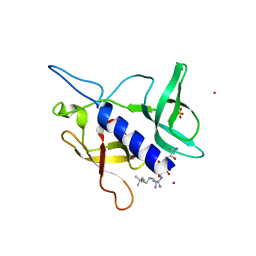 | | Crystal structure of SGF29 tandem tudor domain in complex with a Carba containing peptide | | Descriptor: | Carba-containing peptide, GLYCEROL, SAGA-associated factor 29 homolog, ... | | Authors: | Dong, A, Xu, C, Tempel, W, Cerovina, T, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2015-06-12 | | Release date: | 2015-07-01 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Chemical basis for the recognition of trimethyllysine by epigenetic reader proteins.
Nat Commun, 6, 2015
|
|
3PMT
 
 | | Crystal structure of the Tudor domain of human Tudor domain-containing protein 3 | | Descriptor: | TETRAETHYLENE GLYCOL, Tudor domain-containing protein 3 | | Authors: | Lam, R, Bian, C.B, Guo, Y.H, Xu, C, Kania, J, Bountra, C, Weigelt, J, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-11-18 | | Release date: | 2010-12-01 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of TDRD3 and Methyl-Arginine Binding Characterization of TDRD3, SMN and SPF30.
Plos One, 7, 2012
|
|
8GXP
 
 | | Complex structure of RORgama with betulinic acid | | Descriptor: | Betulinic acid, Nuclear receptor ROR-gamma | | Authors: | Zhang, X.L, Xu, C, Bai, F. | | Deposit date: | 2022-09-20 | | Release date: | 2023-06-07 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Discovery, structural optimization, and anti-tumor bioactivity evaluations of betulinic acid derivatives as a new type of ROR gamma antagonists.
Eur.J.Med.Chem., 257, 2023
|
|
5VC9
 
 | | Zinc finger of human CXXC4 in complex with CpG DNA | | Descriptor: | CXXC-type zinc finger protein 4, CpG DNA, UNKNOWN ATOM OR ION, ... | | Authors: | Liu, K, Xu, C, Tempel, W, Walker, J.R, Arrowsmith, C.H, Bountra, C, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2017-03-31 | | Release date: | 2017-06-28 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | DNA Sequence Recognition of Human CXXC Domains and Their Structural Determinants.
Structure, 26, 2018
|
|
8I3F
 
 | |
4L36
 
 | | Crystal structure of the cytochrome P450 enzyme TxtE | | Descriptor: | HEME B/C, IMIDAZOLE, Putative P450-like protein | | Authors: | Yu, F, Li, M.J, Xu, C.Y, Wang, Z.J, Zhou, H, Yang, M, Chen, Y.X, Tang, L, He, J.H. | | Deposit date: | 2013-06-05 | | Release date: | 2013-12-11 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.104 Å) | | Cite: | Structural Insights into the Mechanism for Recognizing Substrate of the Cytochrome P450 Enzyme TxtE.
Plos One, 8, 2013
|
|
3J89
 
 | | Structural Plasticity of Helical Nanotubes Based on Coiled-Coil Assemblies | | Descriptor: | synthetic peptide | | Authors: | Egelman, E.H, Xu, C, DiMaio, F, Magnotti, E, Modlin, C, Yu, X, Wright, E, Baker, D, Conticello, V.P. | | Deposit date: | 2014-10-07 | | Release date: | 2015-02-11 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural plasticity of helical nanotubes based on coiled-coil assemblies.
Structure, 23, 2015
|
|
8I3G
 
 | | Crystal structure of Eaf3-Eaf7 complex | | Descriptor: | Chromatin modification-related protein EAF3, Chromatin modification-related protein EAF7 | | Authors: | Chen, Z, Xu, C. | | Deposit date: | 2023-01-17 | | Release date: | 2023-05-31 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Molecular basis for Eaf3-mediated assembly of Rpd3S and NuA4.
Cell Discov, 9, 2023
|
|
1YOP
 
 | | The solution structure of Kti11p | | Descriptor: | Kti11p, ZINC ION | | Authors: | Sun, J, Zhang, J, Wu, F, Xu, C, Li, S, Zhao, W, Wu, Z, Wu, J, Zhou, C.-Z, Shi, Y. | | Deposit date: | 2005-01-28 | | Release date: | 2005-04-05 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of Kti11p from Saccharomyces cerevisiae reveals a novel zinc-binding module.
Biochemistry, 44, 2005
|
|
5ZN9
 
 | | Crystal structure of PX domain | | Descriptor: | SULFATE ION, Sorting nexin-27 | | Authors: | Li, Y, Zhu, Z, Li, F, Liao, S, Xu, C. | | Deposit date: | 2018-04-08 | | Release date: | 2019-04-10 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.776 Å) | | Cite: | Crystal structure of PX domain
To Be Published
|
|
8WQI
 
 | |
6ICV
 
 | | Structure of SETD3 bound to SAH and unmodified actin | | Descriptor: | Actin, cytoplasmic 1, Histone-lysine N-methyltransferase setd3, ... | | Authors: | Guo, Q, Liao, S, Xu, C, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-09-07 | | Release date: | 2019-02-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural insights into SETD3-mediated histidine methylation on beta-actin.
Elife, 8, 2019
|
|
6MCP
 
 | | L. pneumophila effector kinase LegK7 (AMP-PNP bound) in complex with human MOB1A | | Descriptor: | DI(HYDROXYETHYL)ETHER, HEXAETHYLENE GLYCOL, LegK7, ... | | Authors: | Beyrakhova, K.A, Xu, C, Boniecki, M.T, Cygler, M. | | Deposit date: | 2018-09-01 | | Release date: | 2019-09-04 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | TheLegionellakinase LegK7 exploits the Hippo pathway scaffold protein MOB1A for allostery and substrate phosphorylation.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
2LLK
 
 | | Solution NMR structure of the N-terminal myb-like 1 domain of the human cyclin-D-binding transcription factor 1 (hDMP1), Northeast Structural Genomics Consortium (NESG) target ID hr8011a | | Descriptor: | Cyclin-D-binding Myb-like transcription factor 1 | | Authors: | Montecchio, M, Lemak, A, Yee, A, Xu, C, Garcia, M, Houliston, S, Min, J, Bellanda, M, Montelione, G.T, Arrowsmith, C, Northeast Structural Genomics Consortium (NESG), Structural Genomics Consortium (SGC) | | Deposit date: | 2011-11-10 | | Release date: | 2011-11-23 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure of the N-terminal myb-like 1 domain of the human cyclin D binding transcription factor 1 (hDMP1).
To be Published
|
|
6J9H
 
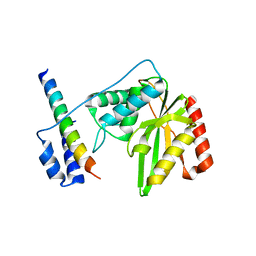 | |
6J8N
 
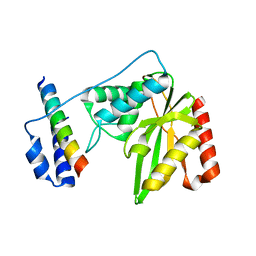 | | Crystal structure of SVBP-VASH1 complex, mutation C169A of VASH1 | | Descriptor: | Small vasohibin-binding protein, Tubulinyl-Tyr carboxypeptidase 1 | | Authors: | Liao, S, Gao, J, Xu, C, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-01-20 | | Release date: | 2019-06-19 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Molecular basis of vasohibins-mediated detyrosination and its impact on spindle function and mitosis.
Cell Res., 29, 2019
|
|
3MEA
 
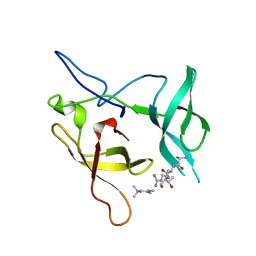 | | Crystal structure of the SGF29 in complex with H3K4me3 | | Descriptor: | Histone H3, SAGA-associated factor 29 homolog | | Authors: | Bian, C, Xu, C, Tempel, W, MacKenzie, F, Bountra, C, Weigelt, J, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-03-31 | | Release date: | 2010-04-28 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.26 Å) | | Cite: | Sgf29 binds histone H3K4me2/3 and is required for SAGA complex recruitment and histone H3 acetylation.
Embo J., 30, 2011
|
|
1R1G
 
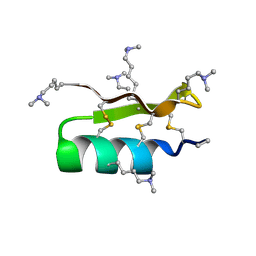 | |
