8KGT
 
 | | Structure of African swine fever virus topoisomerase II in complex with dsDNA | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, DNA topoisomerase 2 | | Authors: | Cong, J, Xin, Y, Li, X, Chen, Y. | | Deposit date: | 2023-08-19 | | Release date: | 2024-04-03 | | Last modified: | 2024-08-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural insights into the DNA topoisomerase II of the African swine fever virus.
Nat Commun, 15, 2024
|
|
8KGS
 
 | | Structure of African swine fever virus topoisomerase II in complex with dsDNA | | Descriptor: | DNA topoisomerase 2, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER | | Authors: | Cong, J, Xin, Y, Li, X, Chen, Y. | | Deposit date: | 2023-08-19 | | Release date: | 2024-04-03 | | Last modified: | 2024-08-14 | | Method: | X-RAY DIFFRACTION (2.603 Å) | | Cite: | Structural insights into the DNA topoisomerase II of the African swine fever virus.
Nat Commun, 15, 2024
|
|
8KGL
 
 | |
8KGO
 
 | |
8KGM
 
 | | Structure of African swine fever virus topoisomerase II in complex with dsDNA | | Descriptor: | DNA (38-MER), DNA topoisomerase 2 | | Authors: | Cong, J, Xin, Y, Li, X, Chen, Y. | | Deposit date: | 2023-08-19 | | Release date: | 2024-04-03 | | Last modified: | 2024-08-14 | | Method: | ELECTRON MICROSCOPY (4.8 Å) | | Cite: | Structural insights into the DNA topoisomerase II of the African swine fever virus.
Nat Commun, 15, 2024
|
|
8KGP
 
 | |
8KGQ
 
 | | Structure of African swine fever virus topoisomerase II in complex with dsDNA | | Descriptor: | DNA (38-MER), DNA topoisomerase 2 | | Authors: | Cong, J, Xin, Y, Li, X, Chen, Y. | | Deposit date: | 2023-08-19 | | Release date: | 2024-04-03 | | Last modified: | 2024-08-14 | | Method: | ELECTRON MICROSCOPY (5.6 Å) | | Cite: | Structural insights into the DNA topoisomerase II of the African swine fever virus.
Nat Commun, 15, 2024
|
|
7WOL
 
 | | Crystal structure of lipase TrLipB from Thermomocrobium roseum | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Zhang, L, Fang, Y, Shi, Y, Gu, Z, Xin, Y. | | Deposit date: | 2022-01-21 | | Release date: | 2023-01-25 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of lipase TrLipB from Thermomocrobium roseum
To Be Published
|
|
6KNE
 
 | |
6KND
 
 | |
7SOY
 
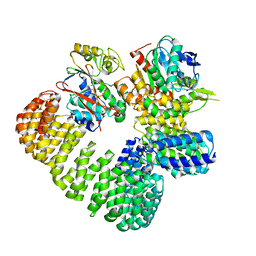 | | The structure of the PP2A-B56gamma1 holoenzyme-PME-1 complex | | Descriptor: | Isoform Gamma-1 of Serine/threonine-protein phosphatase 2A 56 kDa regulatory subunit gamma isoform, Protein phosphatase methylesterase 1, Serine/threonine-protein phosphatase 2A 65 kDa regulatory subunit A alpha isoform, ... | | Authors: | Li, Y, Balakrishnan, V.K, Rowse, M, Novikova, I.V, Xing, Y. | | Deposit date: | 2021-11-01 | | Release date: | 2022-08-31 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Coupling to short linear motifs creates versatile PME-1 activities in PP2A holoenzyme demethylation and inhibition.
Elife, 11, 2022
|
|
1EB6
 
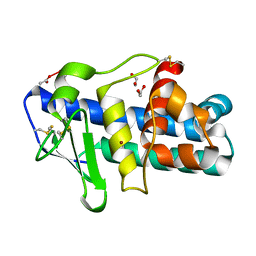 | | Deuterolysin from Aspergillus oryzae | | Descriptor: | 1,2-ETHANEDIOL, NEUTRAL PROTEASE II, ZINC ION | | Authors: | McAuley, K.E, Jia-Xing, Y, Dodson, E.J, Lehmbeck, J, Ostergaard, P.R, Wilson, K.S. | | Deposit date: | 2001-07-19 | | Release date: | 2001-11-23 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | A Quick Solution: Ab Initio Structure Determination of a 19 kDa Metalloproteinase Using Acorn
Acta Crystallogr.,Sect.D, 57, 2001
|
|
4QMG
 
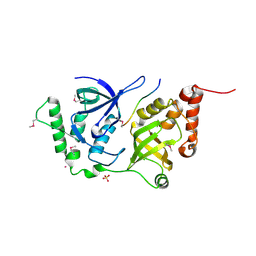 | | The Structure of MTDH-SND1 Complex Reveals Novel Cancer-Promoting Interactions | | Descriptor: | CESIUM ION, GLYCEROL, Protein LYRIC, ... | | Authors: | Guo, F, Stanevich, V, Wan, L, Satyshur, K, Kang, Y, Xing, Y. | | Deposit date: | 2014-06-16 | | Release date: | 2014-10-08 | | Method: | X-RAY DIFFRACTION (2.701 Å) | | Cite: | Structural Insights into the Tumor-Promoting Function of the MTDH-SND1 Complex.
Cell Rep, 8, 2014
|
|
5V0L
 
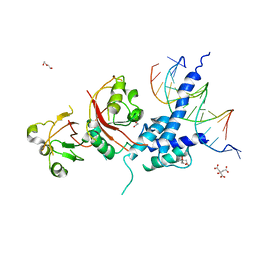 | | Crystal structure of the AHR-ARNT heterodimer in complex with the DRE | | Descriptor: | Aryl hydrocarbon receptor, Aryl hydrocarbon receptor nuclear translocator, CITRIC ACID, ... | | Authors: | Seok, S.-H, Lee, W, Jiang, L, Bradfield, C.A, Xing, Y. | | Deposit date: | 2017-02-28 | | Release date: | 2017-04-19 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (4 Å) | | Cite: | Structural hierarchy controlling dimerization and target DNA recognition in the AHR transcriptional complex.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5W0X
 
 | | Crystal structure of mouse TOR signaling pathway regulator-like (TIPRL) delta 94-103 | | Descriptor: | TIP41-like protein | | Authors: | Wu, C, Zheng, A, Li, J, Satyshur, K, Xing, Y. | | Deposit date: | 2017-06-01 | | Release date: | 2018-01-17 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.717 Å) | | Cite: | Methylation-regulated decommissioning of multimeric PP2A complexes.
Nat Commun, 8, 2017
|
|
5W0W
 
 | | Crystal structure of Protein Phosphatase 2A bound to TIPRL | | Descriptor: | MANGANESE (II) ION, Serine/threonine-protein phosphatase 2A 65 kDa regulatory subunit A alpha isoform, Serine/threonine-protein phosphatase 2A catalytic subunit alpha isoform, ... | | Authors: | Wu, C, Zheng, A, Li, J, Satyshur, K, Xing, Y. | | Deposit date: | 2017-06-01 | | Release date: | 2018-01-17 | | Last modified: | 2020-01-01 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Methylation-regulated decommissioning of multimeric PP2A complexes.
Nat Commun, 8, 2017
|
|
8RIJ
 
 | | Discovery of the first orally bioavailable ADAMTS7 inhibitor BAY-9835 | | Descriptor: | CALCIUM ION, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Schafer, M, Meibom, D, Wasnaire, P, Beyer, K, Broehl, A, Cancho-Grande, Y, Elowe, N, Henninger, K, Johannes, S, Jungmann, N, Krainz, T, Lindner, N, Maassen, S, MacDonald, B, Menshykau, D, Mittendorf, J, Sanchez, G, Stefan, E, Torge, A, Xing, Y, Zubov, D. | | Deposit date: | 2023-12-18 | | Release date: | 2024-02-21 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | BAY-9835: Discovery of the First Orally Bioavailable ADAMTS7 Inhibitor.
J.Med.Chem., 67, 2024
|
|
4JGV
 
 | | Crystal Structure of Human Nur77 Ligand-binding Domain in Complex with THPN | | Descriptor: | 1-(3,4,5-trihydroxyphenyl)nonan-1-one, GLYCEROL, Nuclear receptor subfamily 4 group A member 1 | | Authors: | Zhang, Q, Li, F, Li, A, Tian, X, Wan, W, Wan, Y, Chen, H, Xing, Y, Wu, Q, Lin, T. | | Deposit date: | 2013-03-04 | | Release date: | 2013-12-18 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3.01 Å) | | Cite: | Orphan nuclear receptor TR3 acts in autophagic cell death via mitochondrial signaling pathway.
Nat.Chem.Biol., 10, 2014
|
|
4KZJ
 
 | | Crystal Structure of TR3 LBD L449W Mutant | | Descriptor: | GLYCEROL, Nuclear receptor subfamily 4 group A member 1 | | Authors: | Li, F, Zhang, Q, Li, A, Tian, X, Cai, Q, Wang, W, Wang, Y, Chen, H, Xing, Y, Wu, Q, Lin, T. | | Deposit date: | 2013-05-30 | | Release date: | 2013-12-18 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Orphan nuclear receptor TR3 acts in autophagic cell death via mitochondrial signaling pathway.
Nat.Chem.Biol., 10, 2014
|
|
4KZI
 
 | | Crystal Structure of TR3 LBD in complex with DPDO | | Descriptor: | 1-(3,5-dimethoxyphenyl)decan-1-one, GLYCEROL, Nuclear receptor subfamily 4 group A member 1 | | Authors: | Li, F, Zhang, Q, Li, A, Tian, X, Cai, Q, Wang, W, Wang, Y, Chen, H, Xing, Y, Wu, Q, Lin, T. | | Deposit date: | 2013-05-30 | | Release date: | 2013-12-18 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | Orphan nuclear receptor TR3 acts in autophagic cell death via mitochondrial signaling pathway.
Nat.Chem.Biol., 10, 2014
|
|
4KZM
 
 | | Crystal Structure of TR3 LBD S553A Mutant | | Descriptor: | GLYCEROL, Nuclear receptor subfamily 4 group A member 1 | | Authors: | Li, F, Zhang, Q, Li, A, Tian, X, Cai, Q, Wang, W, Wang, Y, Chen, H, Xing, Y, Wu, Q, Lin, T. | | Deposit date: | 2013-05-30 | | Release date: | 2013-12-18 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Orphan nuclear receptor TR3 acts in autophagic cell death via mitochondrial signaling pathway.
Nat.Chem.Biol., 10, 2014
|
|
3ML6
 
 | | a complex between Dishevelled2 and clathrin adaptor AP-2 | | Descriptor: | Chimeric complex between protein Dishevelled2 homolog dvl-2 and clathrin adaptor AP-2 complex subunit mu | | Authors: | Yu, A, Xing, Y, Harrison, S.C, Kirchhausen, T.L. | | Deposit date: | 2010-04-16 | | Release date: | 2010-08-11 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structural analysis of the interaction between Dishevelled2 and clathrin AP-2 adaptor, a critical step in noncanonical Wnt signaling.
Structure, 18, 2010
|
|
4GD9
 
 | | Circular Permuted Streptavidin N49/G48 | | Descriptor: | BIOTIN, SULFATE ION, Streptavidin | | Authors: | Le Trong, I, Chu, V, Xing, Y, Lybrand, T.P, Stayton, P.S, Stenkamp, R.E. | | Deposit date: | 2012-07-31 | | Release date: | 2013-06-05 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural consequences of cutting a binding loop: two circularly permuted variants of streptavidin.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
8U1X
 
 | | The structure of the PP2A-B56Delta holoenzyme mutant - E197K | | Descriptor: | MANGANESE (II) ION, Serine/threonine-protein phosphatase 2A 56 kDa regulatory subunit delta isoform, Serine/threonine-protein phosphatase 2A 65 kDa regulatory subunit A alpha isoform, ... | | Authors: | Wu, C.G, Xing, Y. | | Deposit date: | 2023-09-04 | | Release date: | 2024-01-17 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | B56 delta long-disordered arms form a dynamic PP2A regulation interface coupled with global allostery and Jordan's syndrome mutations.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
4GDA
 
 | | Circular Permuted Streptavidin A50/N49 | | Descriptor: | BIOTIN, ETHANOL, GLYCEROL, ... | | Authors: | Le Trong, I, Chu, V, Xing, Y, Lybrand, T.P, Stayton, P.S, Stenkamp, R.E. | | Deposit date: | 2012-07-31 | | Release date: | 2013-06-05 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Structural consequences of cutting a binding loop: two circularly permuted variants of streptavidin.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
