8WOT
 
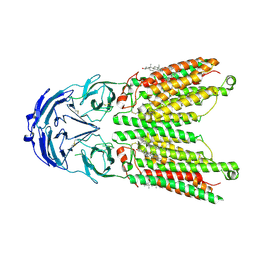 | | Cryo-EM structure of human SIDT1 protein with C2 symmetry at low pH | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHOLESTEROL, ... | | Authors: | Liu, W, Tang, M, Wang, J, Zhang, X, Wu, S, Ru, H. | | Deposit date: | 2023-10-07 | | Release date: | 2024-03-20 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.18 Å) | | Cite: | Structural insights into cholesterol transport and hydrolase activity of a putative human RNA transport protein SIDT1.
Cell Discov, 10, 2024
|
|
8WOQ
 
 | | Cryo-EM structure of human SIDT1 protein with C1 symmetry at neutral pH | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Liu, W, Tang, M, Wang, J, Zhang, X, Wu, S, Ru, H. | | Deposit date: | 2023-10-07 | | Release date: | 2024-03-20 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.85 Å) | | Cite: | Structural insights into cholesterol transport and hydrolase activity of a putative human RNA transport protein SIDT1.
Cell Discov, 10, 2024
|
|
8WOS
 
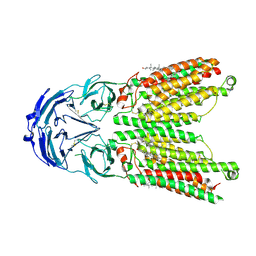 | | Cryo-EM structure of human SIDT1 protein with C1 symmetry at low pH | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHOLESTEROL, ... | | Authors: | Liu, W, Tang, M, Wang, J, Zhang, X, Wu, S, Ru, H. | | Deposit date: | 2023-10-07 | | Release date: | 2024-03-20 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.37 Å) | | Cite: | Structural insights into cholesterol transport and hydrolase activity of a putative human RNA transport protein SIDT1.
Cell Discov, 10, 2024
|
|
3JCD
 
 | | Structure of Escherichia coli EF4 in posttranslocational ribosomes (Post EF4) | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Zhang, D, Yan, K, Liu, G, Song, G, Luo, J, Shi, Y, Cheng, E, Wu, S, Jiang, T, Low, J, Gao, N, Qin, Y. | | Deposit date: | 2015-12-01 | | Release date: | 2016-01-13 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | EF4 disengages the peptidyl-tRNA CCA end and facilitates back-translocation on the 70S ribosome
Nat. Struct. Mol. Biol., 23, 2016
|
|
3JCE
 
 | | Structure of Escherichia coli EF4 in pretranslocational ribosomes (Pre EF4) | | Descriptor: | 16S ribosomal RNA, 23 ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Zhang, D, Yan, K, Liu, G, Song, G, Luo, J, Shi, Y, Cheng, E, Wu, S, Jiang, T, Low, J, Gao, N, Qin, Y. | | Deposit date: | 2015-12-01 | | Release date: | 2016-01-13 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | EF4 disengages the peptidyl-tRNA CCA end and facilitates back-translocation on the 70S ribosome
Nat. Struct. Mol. Biol., 23, 2016
|
|
3J95
 
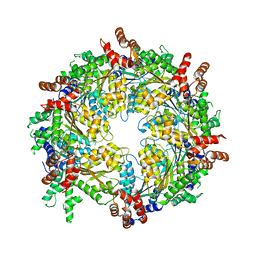 | | Structure of ADP-bound N-ethylmaleimide sensitive factor determined by single particle cryoelectron microscopy | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Vesicle-fusing ATPase | | Authors: | Zhao, M, Wu, S, Cheng, Y, Brunger, A.T. | | Deposit date: | 2014-12-05 | | Release date: | 2015-01-28 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (7.6 Å) | | Cite: | Mechanistic insights into the recycling machine of the SNARE complex.
Nature, 518, 2015
|
|
3J94
 
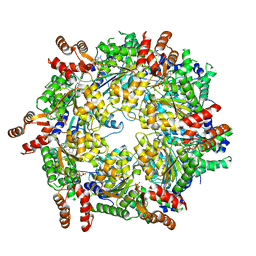 | | Structure of ATP-bound N-ethylmaleimide sensitive factor determined by single particle cryoelectron microscopy | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Vesicle-fusing ATPase | | Authors: | Zhao, M, Wu, S, Cheng, Y, Brunger, A.T. | | Deposit date: | 2014-12-05 | | Release date: | 2015-01-28 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Mechanistic insights into the recycling machine of the SNARE complex.
Nature, 518, 2015
|
|
1W60
 
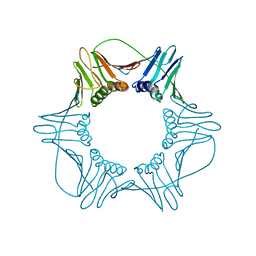 | | NATIVE HUMAN PCNA | | Descriptor: | PROLIFERATING CELL NUCLEAR ANTIGEN | | Authors: | Kontopidis, G, Wu, S, Zheleva, D, Taylor, P, Mcinnes, C, Lane, D, Fischer, P, Walkinshaw, M. | | Deposit date: | 2004-08-11 | | Release date: | 2005-01-13 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Structural and Biochemical Studies of Human Proliferating Cell Nuclear Antigen Complexes Provide a Rationale for Cyclin Association and Inhibitor Design
Proc.Natl.Acad.Sci.USA, 102, 2005
|
|
7VT8
 
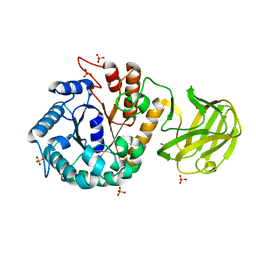 | | Crystal structure of MtGlu5 from Meiothermus taiwanensis WR-220 | | Descriptor: | Endoglucanase H, SULFATE ION, beta-D-glucopyranose | | Authors: | Ye, T.J, Ko, P.T, Huang, K.F, Wu, S.H. | | Deposit date: | 2021-10-28 | | Release date: | 2022-09-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.99 Å) | | Cite: | Synergic action of an inserted carbohydrate-binding module in a glycoside hydrolase family 5 endoglucanase.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
1VYJ
 
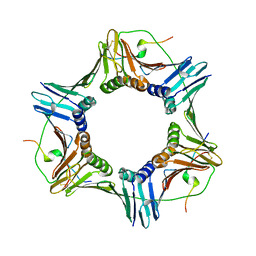 | | Structural and biochemical studies of human PCNA complexes provide the basis for association with CDK/cyclin and rationale for inhibitor design | | Descriptor: | PROLIFERATING CELL NUCLEAR ANTIGEN, SMALL PEPTIDE SAVLQKKITDYFHPKK | | Authors: | Kontopidis, G, Wu, S, Zheleva, D, Taylor, P, Mcinnes, C, Lane, D, Fischer, P, Walkinshaw, M.D. | | Deposit date: | 2004-04-30 | | Release date: | 2005-01-13 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural and Biochemical Studies of Human Proliferating Cell Nuclear Antigen Complexes Provide a Rationale for Cyclin Association and Inhibitor Design
Proc.Natl.Acad.Sci.USA, 102, 2005
|
|
3J96
 
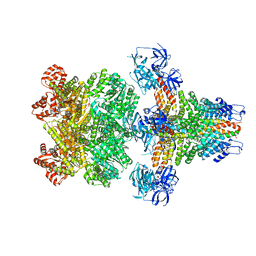 | | Structure of 20S supercomplex determined by single particle cryoelectron microscopy (State I) | | Descriptor: | Alpha-soluble NSF attachment protein, Synaptosomal-associated protein 25, Syntaxin-1A, ... | | Authors: | Zhao, M, Wu, S, Cheng, Y, Brunger, A.T. | | Deposit date: | 2014-12-05 | | Release date: | 2015-01-28 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (7.6 Å) | | Cite: | Mechanistic insights into the recycling machine of the SNARE complex.
Nature, 518, 2015
|
|
5IHW
 
 | | The crystal structure of SdrE from staphylococcus aureus | | Descriptor: | Serine-aspartate repeat-containing protein E | | Authors: | Zhang, S, Wei, J, Wu, S, Zhang, X, Luo, M, Wang, D. | | Deposit date: | 2016-02-29 | | Release date: | 2017-03-22 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | The crystal structure of SdrE from staphylococcus aureus
To Be Published
|
|
3J99
 
 | | Structure of 20S supercomplex determined by single particle cryoelectron microscopy (State IIIb) | | Descriptor: | Alpha-soluble NSF attachment protein, Synaptosomal-associated protein 25, Syntaxin-1A, ... | | Authors: | Zhao, M, Wu, S, Cheng, Y, Brunger, A.T. | | Deposit date: | 2014-12-05 | | Release date: | 2015-01-28 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (8.2 Å) | | Cite: | Mechanistic insights into the recycling machine of the SNARE complex.
Nature, 518, 2015
|
|
5WUG
 
 | | Expression, characterization and crystal structure of a novel beta-glucosidase from Paenibacillus barengoltzii | | Descriptor: | Beta-glucosidase | | Authors: | Jiang, Z, Wu, S, Yang, D, Qin, Z, You, X, Huang, P. | | Deposit date: | 2016-12-17 | | Release date: | 2018-01-31 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.216 Å) | | Cite: | Expression, Biochemical Characterization and Structure Resolution of beta-glucosidase from Paenibacillus barengoltzii
J Food Sci Technol(China), 2019
|
|
3J98
 
 | | Structure of 20S supercomplex determined by single particle cryoelectron microscopy (State IIIa) | | Descriptor: | Alpha-soluble NSF attachment protein, Synaptosomal-associated protein 25, Syntaxin-1A, ... | | Authors: | Zhao, M, Wu, S, Cheng, Y, Brunger, A.T. | | Deposit date: | 2014-12-05 | | Release date: | 2015-01-28 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (8.4 Å) | | Cite: | Mechanistic insights into the recycling machine of the SNARE complex.
Nature, 518, 2015
|
|
1X37
 
 | | Structure of Bacillus subtilis Lon protease SSD domain | | Descriptor: | ATP-dependent protease La 1 | | Authors: | Wang, I, Lou, Y.C, Lo, S.C, Lee, Y.L, Wu, S.H, Chen, C. | | Deposit date: | 2005-04-30 | | Release date: | 2005-10-30 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structural basis and DNA binding property of SSD domain of Bacillus subtilis Lon protease
to be published
|
|
1FKN
 
 | | Structure of Beta-Secretase Complexed with Inhibitor | | Descriptor: | MEMAPSIN 2, inhibitor | | Authors: | Hong, L, Koelsch, G, Lin, X, Wu, S, Terzyan, S, Ghosh, A, Zhang, X.C, Tang, J. | | Deposit date: | 2000-08-09 | | Release date: | 2000-10-09 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of the protease domain of memapsin 2 (beta-secretase) complexed with inhibitor.
Science, 290, 2000
|
|
1Z33
 
 | | Crystal structure of Trichomonas vaginalis purine nucleoside phosphorylase | | Descriptor: | purine nucleoside phosphorylase | | Authors: | Zhang, Y, Wang, W.H, Wu, S.W, Wang, C.C, Ealick, S.E. | | Deposit date: | 2005-03-10 | | Release date: | 2005-03-29 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Identification of a subversive substrate of Trichomonas vaginalis purine nucleoside phosphorylase and the crystal structure of the enzyme-substrate complex.
J.Biol.Chem., 280, 2005
|
|
1Z39
 
 | | Crystal structure of Trichomonas vaginalis purine nucleoside phosphorylase complexed with 2'-deoxyinosine | | Descriptor: | 2'-DEOXYINOSINE, purine nucleoside phosphorylase | | Authors: | Zhang, Y, Wang, W.H, Wu, S.W, Wang, C.C, Ealick, S.E. | | Deposit date: | 2005-03-10 | | Release date: | 2005-03-29 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Identification of a subversive substrate of Trichomonas vaginalis purine nucleoside phosphorylase and the crystal structure of the enzyme-substrate complex.
J.Biol.Chem., 280, 2005
|
|
1Z38
 
 | | Crystal structure of Trichomonas vaginalis purine nucleoside phosphorylase complexed with inosine | | Descriptor: | INOSINE, purine nucleoside phosphorylase | | Authors: | Zhang, Y, Wang, W.H, Wu, S.W, Wang, C.C, Ealick, S.E. | | Deposit date: | 2005-03-10 | | Release date: | 2005-03-29 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Identification of a subversive substrate of Trichomonas vaginalis purine nucleoside phosphorylase and the crystal structure of the enzyme-substrate complex.
J.Biol.Chem., 280, 2005
|
|
1Z35
 
 | | Crystal structure of Trichomonas vaginalis purine nucleoside phosphorylase complexed with 2-fluoroadenosine | | Descriptor: | 2-(6-AMINO-2-FLUORO-PURIN-9-YL)-5-HYDROXYMETHYL-TETRAHYDRO-FURAN-3,4-DIOL, purine nucleoside phosphorylase | | Authors: | Zhang, Y, Wang, W.H, Wu, S.W, Wang, C.C, Ealick, S.E. | | Deposit date: | 2005-03-10 | | Release date: | 2005-03-29 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Identification of a subversive substrate of Trichomonas vaginalis purine nucleoside phosphorylase and the crystal structure of the enzyme-substrate complex.
J.Biol.Chem., 280, 2005
|
|
1PXO
 
 | | HUMAN CYCLIN DEPENDENT KINASE 2 COMPLEXED WITH THE INHIBITOR [4-(2-Amino-4-methyl-thiazol-5-yl)-pyrimidin-2-yl]-(3-nitro-phenyl)-amine | | Descriptor: | Cell division protein kinase 2, [4-(2-AMINO-4-METHYL-THIAZOL-5-YL)-PYRIMIDIN-2-YL]-(3-NITRO-PHENYL)-AMINE | | Authors: | Wang, S, Meades, C, Wood, G, Osnowski, A, Anderson, S, Yuill, R, Thomas, M, Mezna, M, Jackson, W, Midgley, C, Griffiths, G, McNae, I, Wu, S.Y, McInnes, C, Zheleva, D, Walkinshaw, M.D, Fischer, P.M. | | Deposit date: | 2003-07-04 | | Release date: | 2004-04-13 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | 2-Anilino-4-(thiazol-5-yl)pyrimidine CDK inhibitors: synthesis, SAR analysis, X-ray crystallography, and biological activity.
J.Med.Chem., 47, 2004
|
|
1PXM
 
 | | HUMAN CYCLIN DEPENDENT KINASE 2 COMPLEXED WITH THE INHIBITOR 3-[4-(2,4-Dimethyl-thiazol-5-yl)-pyrimidin-2-ylamino]-phenol | | Descriptor: | 3-[4-(2,4-DIMETHYL-THIAZOL-5-YL)-PYRIMIDIN-2-YLAMINO]-PHENOL, Cell division protein kinase 2 | | Authors: | Wang, S, Meades, C, Wood, G, Osnowski, A, Anderson, S, Yuill, R, Thomas, M, Jackson, W, Midgley, C, Griffiths, G, McNae, I, Wu, S.Y, McInnes, C, Zheleva, D, Walkinshaw, M.D, Fischer, P.M. | | Deposit date: | 2003-07-04 | | Release date: | 2004-04-13 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.53 Å) | | Cite: | 2-Anilino-4-(thiazol-5-yl)pyrimidine CDK inhibitors: synthesis, SAR analysis, X-ray crystallography, and biological activity.
J.Med.Chem., 47, 2004
|
|
1Z34
 
 | | Crystal structure of Trichomonas vaginalis purine nucleoside phosphorylase complexed with 2-fluoro-2'-deoxyadenosine | | Descriptor: | 5-(6-AMINO-2-FLUORO-PURIN-9-YL)-2-HYDROXYMETHYL-TETRAHYDRO-FURAN-3-OL, purine nucleoside phosphorylase | | Authors: | Zhang, Y, Wang, W.H, Wu, S.W, Wang, C.C, Ealick, S.E. | | Deposit date: | 2005-03-10 | | Release date: | 2005-03-29 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Identification of a subversive substrate of Trichomonas vaginalis purine nucleoside phosphorylase and the crystal structure of the enzyme-substrate complex.
J.Biol.Chem., 280, 2005
|
|
1PXN
 
 | | HUMAN CYCLIN DEPENDENT KINASE 2 COMPLEXED WITH THE INHIBITOR 4-[4-(4-Methyl-2-methylamino-thiazol-5-yl)-pyrimidin-2-ylamino]-phenol | | Descriptor: | 4-[4-(4-METHYL-2-METHYLAMINO-THIAZOL-5-YL)-PYRIMIDIN-2-YLAMINO]-PHENOL, Cell division protein kinase 2 | | Authors: | Wang, S, Meades, C, Wood, G, Osnowski, A, Anderson, S, Yuill, R, Thomas, M, Mezna, M, Jackson, W, Midgley, C, Griffiths, G, McNae, I, Wu, S.Y, McInnes, C, Zheleva, D, Walkinshaw, M.D, Fischer, P.M. | | Deposit date: | 2003-07-04 | | Release date: | 2004-04-13 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | 2-Anilino-4-(thiazol-5-yl)pyrimidine CDK inhibitors: synthesis, SAR analysis, X-ray crystallography, and biological activity.
J.Med.Chem., 47, 2004
|
|
