3P12
 
 | | Crystal Structure of D-ribose Pyranase Sa240 | | Descriptor: | D-ribose pyranase, GLYCEROL | | Authors: | Wu, M, Wang, L, Zang, J. | | Deposit date: | 2010-09-29 | | Release date: | 2011-05-25 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Crystal structure of Sa240: A ribose pyranase homolog with partial active site from Staphylococcus aureus
J.Struct.Biol., 174, 2011
|
|
3P13
 
 | |
1MWG
 
 | |
1MIS
 
 | |
3K80
 
 | | Structure of essential protein from Trypanosoma brucei | | Descriptor: | Antibody, MP18 RNA editing complex protein | | Authors: | Wu, M. | | Deposit date: | 2009-10-13 | | Release date: | 2010-11-17 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structures of a key interaction protein from the Trypanosoma brucei editosome in complex with single domain antibodies.
J.Struct.Biol., 174, 2011
|
|
3K7U
 
 | | Structure of essential protein from Trypanosoma brucei | | Descriptor: | Antibody, MP18 RNA editing complex protein | | Authors: | Wu, M. | | Deposit date: | 2009-10-13 | | Release date: | 2010-11-17 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structures of a key interaction protein from the Trypanosoma brucei editosome in complex with single domain antibodies.
J.Struct.Biol., 174, 2011
|
|
1YHN
 
 | | Structure basis of RILP recruitment by Rab7 | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Rab interacting lysosomal protein, ... | | Authors: | Wu, M, Wang, T, Hong, W, Song, H. | | Deposit date: | 2005-01-10 | | Release date: | 2005-05-17 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural basis for recruitment of RILP by small GTPase Rab7.
Embo J., 24, 2005
|
|
4KJS
 
 | | Structure of native YfkE | | Descriptor: | cation exchanger YfkE | | Authors: | Wu, M, Tong, S, Zheng, L. | | Deposit date: | 2013-05-03 | | Release date: | 2013-06-26 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Crystal structure of Ca2+/H+ antiporter protein YfkE reveals the mechanisms of Ca2+ efflux and its pH regulation.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4KJR
 
 | |
4E4H
 
 | | Crystal structure of Histone Demethylase NO66 | | Descriptor: | ACETATE ION, DI(HYDROXYETHYL)ETHER, FE (III) ION, ... | | Authors: | Wu, M, Tao, Y, Zang, J. | | Deposit date: | 2012-03-13 | | Release date: | 2013-01-30 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | tructural Basis for Tetramerization-dependent Gene Repression by Histone Demethylase NO66
To be Published
|
|
4GO1
 
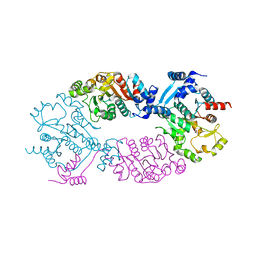 | | Crystal Structure of full length transcription repressor LsrR from E. coli. | | Descriptor: | GLYCEROL, Transcriptional regulator LsrR | | Authors: | Wu, M, Tao, Y, Liu, X, Zang, J. | | Deposit date: | 2012-08-17 | | Release date: | 2013-04-24 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural Basis for Phosphorylated Autoinducer-2 Modulation of the Oligomerization State of the Global Transcription Regulator LsrR from Escherichia coli
J.Biol.Chem., 288, 2013
|
|
4J0C
 
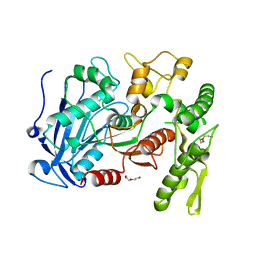 | | tannin acyl hydrolase from Lactobacillus plantarum (native structure) | | Descriptor: | DI(HYDROXYETHYL)ETHER, PENTAETHYLENE GLYCOL, Tannase | | Authors: | Wu, M, Wang, Q, Peng, X, Wen, H, Chen, Q, McKinstry, W.J. | | Deposit date: | 2013-01-30 | | Release date: | 2013-05-22 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal structure of tannase from Lactobacillus plantarum.
J.Mol.Biol., 425, 2013
|
|
4ALC
 
 | | X-Ray photoreduction of Polysaccharide monooxigenase CBM33 | | Descriptor: | CHITIN BINDING PROTEIN, COPPER (II) ION, DI(HYDROXYETHYL)ETHER | | Authors: | Gudmundsson, M, Wu, M, Ishida, T, Momeni, M.H, Vaaje-Kolstad, G, Eijsink, V, Sandgren, M. | | Deposit date: | 2012-03-02 | | Release date: | 2013-02-27 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Structural and Electronic Snapshots During the Transition from a Cu(II) to Cu(I) Metal Center of a Lytic Polysaccharide Monooxygenase by X-Ray Photo-Reduction.
J.Biol.Chem., 289, 2014
|
|
6T8W
 
 | | Complement factor B in complex with (-)-4-(1-((5,7-Dimethyl-1H-indol-4-yl)methyl)piperidin-2-yl)benzoic acid | | Descriptor: | 5,7-dimethyl-4-[[(2~{S})-2-phenylpiperidin-1-yl]methyl]-1~{H}-indole, Complement factor B, SULFATE ION, ... | | Authors: | Mainolfi, N, Ehara, T, Karki, R.G, Anderson, K, Sweeney, A.M, Wiesmann, C, Adams, C, Mainolfi, N, Liao, S.M, Argikar, U.A, Jendza, K, Zhang, C, Powers, J, Klosowski, D.W, Crowley, M, Kawanami, T, Ding, J, April, M, Forster, C, Wu, M.S, Capparelli, M, Ramqaj, R, Solovay, C, Cumin, F, Smith, T.M, Ferrara, L, Lee, W, Long, D, Prentiss, M, Erkenez, A.D, Yang, L, Fang, L, Sellner, H, Sirockin, F, Valeur, E, Erbel, P, Ramage, P, Gerhartz, B, Schubart, A, Flohr, S, Gradoux, N, Feifel, R, Vogg, B, Wiesmann, C, Maibaum, J, Eder, J, Sedrani, R, Harrison, R.A, Mogi, M, Jaffee, B.D, Adams, C.M. | | Deposit date: | 2019-10-25 | | Release date: | 2020-03-04 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Discovery of 4-((2S,4S)-4-Ethoxy-1-((5-methoxy-7-methyl-1H-indol-4-yl)methyl)piperidin-2-yl)benzoic Acid (LNP023), a Factor B Inhibitor Specifically Designed To Be Applicable to Treating a Diverse Array of Complement Mediated Diseases.
J.Med.Chem., 63, 2020
|
|
3C57
 
 | |
5WWL
 
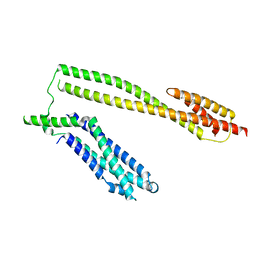 | | Crystal structure of the Schizogenesis pombe kinetochore Mis12C subcomplex | | Descriptor: | Centromere protein mis12, Kinetochore protein nnf1 | | Authors: | Wang, C, Zhou, X, Wu, M, Zhang, X, Zang, J. | | Deposit date: | 2017-01-02 | | Release date: | 2017-11-15 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Phosphorylation of CENP-C by Aurora B facilitates kinetochore attachment error correction in mitosis.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
6KPC
 
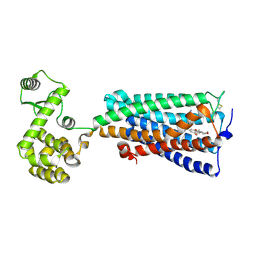 | | Crystal structure of an agonist bound GPCR | | Descriptor: | 7-[(6aR,9R,10aR)-1-Hydroxy-9-(hydroxymethyl)-6,6-dimethyl-6a,7,8,9,10,10a-hexahydro-6H-benzo[c]chromen-3-yl]- 7-methyloctanenitrile, Cannabinoid receptor 2,Endolysin | | Authors: | Li, X.T, Hua, T, Wu, L.J, Makriyannis, A, Wu, M, Liu, Z.J. | | Deposit date: | 2019-08-15 | | Release date: | 2020-02-12 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Activation and Signaling Mechanism Revealed by Cannabinoid Receptor-GiComplex Structures.
Cell, 180, 2020
|
|
6KP6
 
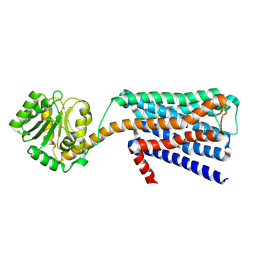 | | The structural study of mutation induced inactivation of Human muscarinic receptor M4 | | Descriptor: | Muscarinic acetylcholine receptor M4,GlgA glycogen synthase,Muscarinic acetylcholine receptor M4 | | Authors: | Wang, J.J, Wu, M, Wu, L.J, Liu, Z.J, Hua, T. | | Deposit date: | 2019-08-14 | | Release date: | 2020-03-11 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The structural study of mutation-induced inactivation of human muscarinic receptor M4
Iucrj, 7, 2020
|
|
7V68
 
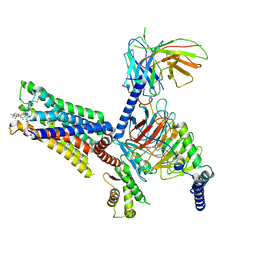 | | An Agonist and PAM-bound Class A GPCR with Gi protein complex structure | | Descriptor: | 3-amino-5-chloro-N-cyclopropyl-4-methyl-6-[2-(4-methylpiperazin-1-yl)-2-oxoethoxy]thieno[2,3-b]pyridine-2-carboxamide, 4-(4,5-dihydro-1,2-oxazol-3-yloxy)-N,N,N-trimethylbut-2-yn-1-aminium, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Wang, J.J, Wu, L.J, Wu, M, Hua, T, Liu, Z.J, Wang, T. | | Deposit date: | 2021-08-20 | | Release date: | 2022-05-11 | | Last modified: | 2022-11-23 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | The unconventional activation of the muscarinic acetylcholine receptor M4R by diverse ligands.
Nat Commun, 13, 2022
|
|
7V6A
 
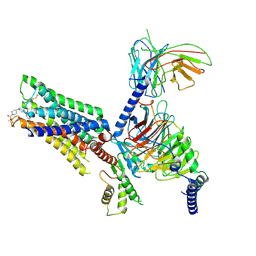 | | Cry-EM structure of M4-c110-G protein complex | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(i) subunit alpha-1, ... | | Authors: | Wang, J.J, Wu, M, Wu, L.J, Hua, T, Liu, Z.J, Wang, T. | | Deposit date: | 2021-08-20 | | Release date: | 2022-05-11 | | Last modified: | 2022-11-23 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | The unconventional activation of the muscarinic acetylcholine receptor M4R by diverse ligands.
Nat Commun, 13, 2022
|
|
7V69
 
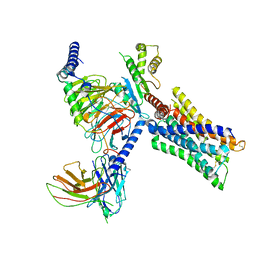 | | Cryo-EM structure of a class A GPCR-G protein complex | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(i) subunit alpha-1, ... | | Authors: | Wang, J.J, Wu, M, Wu, L.J, Hua, T, Liu, Z.J, Wang, T. | | Deposit date: | 2021-08-20 | | Release date: | 2022-05-11 | | Last modified: | 2022-11-23 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | The unconventional activation of the muscarinic acetylcholine receptor M4R by diverse ligands.
Nat Commun, 13, 2022
|
|
3MP1
 
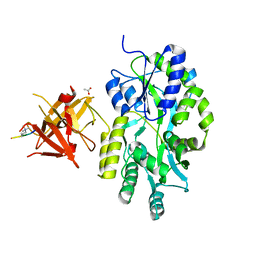 | | Complex structure of Sgf29 and trimethylated H3K4 | | Descriptor: | ACETATE ION, H3K4me3 peptide, Maltose-binding periplasmic protein,LINKER,SAGA-associated factor 29, ... | | Authors: | Li, J, Ruan, J, Wu, M, Xue, X, Zang, J. | | Deposit date: | 2010-04-24 | | Release date: | 2011-05-04 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Sgf29 binds histone H3K4me2/3 and is required for SAGA complex recruitment and histone H3 acetylation
Embo J., 30, 2011
|
|
3MP6
 
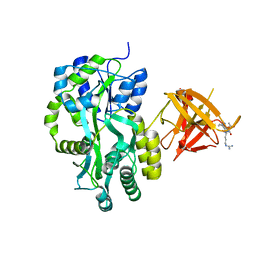 | | Complex Structure of Sgf29 and dimethylated H3K4 | | Descriptor: | H3K4me2 peptide, Maltose-binding periplasmic protein,LINKER,SAGA-associated factor 29, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Li, J, Wu, M, Ruan, J, Zang, J. | | Deposit date: | 2010-04-25 | | Release date: | 2011-05-04 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Sgf29 binds histone H3K4me2/3 and is required for SAGA complex recruitment and histone H3 acetylation
Embo J., 30, 2011
|
|
3MP8
 
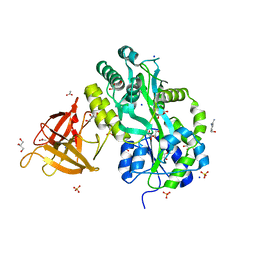 | | Crystal structure of Sgf29 tudor domain | | Descriptor: | 4-(HYDROXYMETHYL)BENZAMIDINE, ACETIC ACID, GLYCEROL, ... | | Authors: | Li, J, Wu, M, Ruan, J, Zang, J. | | Deposit date: | 2010-04-26 | | Release date: | 2011-05-04 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Sgf29 binds histone H3K4me2/3 and is required for SAGA complex recruitment and histone H3 acetylation
Embo J., 30, 2011
|
|
6XRZ
 
 | | The 28-kDa Frameshift Stimulation Element from the SARS-CoV-2 RNA Genome | | Descriptor: | Frameshift Stimulation Element from the SARS-CoV-2 RNA Genome | | Authors: | Zhang, K, Zheludev, I, Hagey, R, Wu, M, Haslecker, R, Hou, Y, Kretsch, R, Pintilie, G, Rangan, R, Kladwang, W, Li, S, Pham, E, Souibgui, C, Baric, R, Sheahan, T, Souza, V, Glenn, J, Chiu, W, Das, R. | | Deposit date: | 2020-07-14 | | Release date: | 2020-08-19 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (6.9 Å) | | Cite: | Cryo-electron Microscopy and Exploratory Antisense Targeting of the 28-kDa Frameshift Stimulation Element from the SARS-CoV-2 RNA Genome.
Biorxiv, 2020
|
|
