8WQY
 
 | | Fe-O nanocluster of form-XI in the 4-fold channel of Ureaplasma diversum ferritin | | Descriptor: | FE (III) ION, ferritin | | Authors: | Wang, W.M, Ma, D.Y, Gong, W.J, Wu, L.J, Wang, H.F. | | Deposit date: | 2023-10-12 | | Release date: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Growth Process of Fe-O Nanoclusters with Different Sizes Biosynthesized by Protein Nanocages.
J.Am.Chem.Soc., 146, 2024
|
|
1YJ0
 
 | | Crystal Structures of Chicken Annexin V in Complex with Zn2+ | | Descriptor: | Annexin A5, SULFATE ION, ZINC ION | | Authors: | Ortlund, E.A, Chai, G, Genge, B, Wu, L.N.Y, Wuthier, R.E, Lebioda, L. | | Deposit date: | 2005-01-11 | | Release date: | 2005-03-08 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Crystal Structures of Chicken Annexin A5 in Complex with Functional Modifiers Ca2+ and Zn2+ Reveal Zn2+ Induced Formation of Non-Planar Assemblies
Annexins, 1, 2005
|
|
8HRD
 
 | | Crystal structure of the receptor binding domain of SARS-CoV-2 Delta variant in complex with IMCAS74 Fab and W14 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, IMCAS74 Fab heavy chain, IMCAS74 Fab light chain, ... | | Authors: | Zhao, R.C, Wu, L.L, Han, P. | | Deposit date: | 2022-12-15 | | Release date: | 2023-12-20 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.86 Å) | | Cite: | Defining a de novo non-RBM antibody as RBD-8 and its synergistic rescue of immune-evaded antibodies to neutralize Omicron SARS-CoV-2.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
1YII
 
 | | Crystal Structures of Chicken Annexin V in Complex with Ca2+ | | Descriptor: | Annexin A5, CALCIUM ION, SULFATE ION | | Authors: | Ortlund, E.A, Chai, G, Genge, B, Wu, L.N.Y, Wuthier, R.E, Lebioda, L. | | Deposit date: | 2005-01-11 | | Release date: | 2005-03-08 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Crystal Structures of Chicken Annexin A5 in Complex with Functional Modifiers Ca2+ and Zn2+ Reveal Zn2+ Induced Formation of Non-Planar Assemblies
Annexins, 1, 2005
|
|
4DGX
 
 | | LEOPARD Syndrome-Associated SHP2/Y279C mutant | | Descriptor: | Tyrosine-protein phosphatase non-receptor type 11 | | Authors: | Yu, Z.H, Xu, J, Walls, C.D, Chen, L, Zhang, S, Wu, L, Wang, L.N, Liu, S.J, Zhang, Z.Y. | | Deposit date: | 2012-01-27 | | Release date: | 2013-03-06 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural and Mechanistic Insights into LEOPARD Syndrome-Associated SHP2 Mutations.
J.Biol.Chem., 288, 2013
|
|
8GPT
 
 | | YFV_E_YD6scFv_postfusion | | Descriptor: | Envelope protein, YD6_VH, YD6_VL | | Authors: | Li, Y, Wu, L, Qi, J, Yan, J, Gao, G.F. | | Deposit date: | 2022-08-27 | | Release date: | 2022-11-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.07 Å) | | Cite: | A neutralizing-protective supersite of human monoclonal antibodies for yellow fever virus.
Innovation (N Y), 3, 2022
|
|
4DGP
 
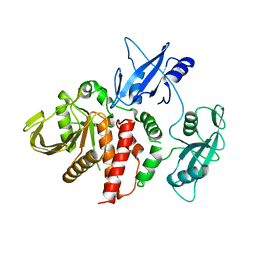 | | The wild-type Src homology 2 (SH2)-domain containing protein tyrosine phosphatase-2 (SHP2) | | Descriptor: | Tyrosine-protein phosphatase non-receptor type 11 | | Authors: | Yu, Z.H, Xu, J, Walls, C.D, Chen, L, Zhang, S, Wu, L, Wang, L.N, Liu, S.J, Zhang, Z.Y. | | Deposit date: | 2012-01-26 | | Release date: | 2013-03-06 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural and Mechanistic Insights into LEOPARD Syndrome-Associated SHP2 Mutations.
J.Biol.Chem., 288, 2013
|
|
7DL0
 
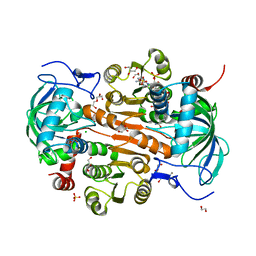 | | The mutant E310G/A314Y of 3,5-DAHDHcca complex with NADPH | | Descriptor: | 1,2-ETHANEDIOL, 3,5-diaminohexanoate dehydrogenase, CHLORIDE ION, ... | | Authors: | Liu, N, Wu, L, Zhu, D.M, Zhou, J.H. | | Deposit date: | 2020-11-25 | | Release date: | 2021-09-29 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Crystal Structures and Catalytic Mechanism of l-erythro-3,5-Diaminohexanoate Dehydrogenase and Rational Engineering for Asymmetric Synthesis of beta-Amino Acids.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
7DL7
 
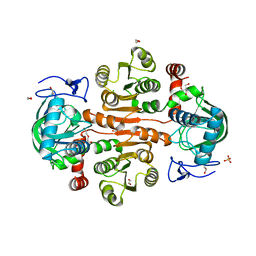 | | The wild-type structure of 3,5-DAHDHcca | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 3,5-diaminohexanoate dehydrogenase, ... | | Authors: | Liu, N, Wu, L, Zhu, D.M, Zhou, J.H. | | Deposit date: | 2020-11-26 | | Release date: | 2021-09-29 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.30065823 Å) | | Cite: | Crystal Structures and Catalytic Mechanism of l-erythro-3,5-Diaminohexanoate Dehydrogenase and Rational Engineering for Asymmetric Synthesis of beta-Amino Acids.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
7DL3
 
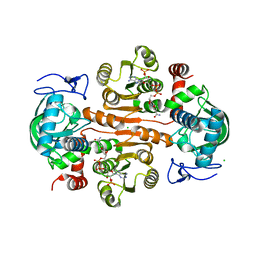 | | The structure of 3,5-DAHDHcca complex with NADPH | | Descriptor: | 3,5-diaminohexanoate dehydrogenase, CHLORIDE ION, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Liu, N, Wu, L, Zhu, D.M, Zhou, J.H. | | Deposit date: | 2020-11-25 | | Release date: | 2021-09-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.84606934 Å) | | Cite: | Crystal Structures and Catalytic Mechanism of l-erythro-3,5-Diaminohexanoate Dehydrogenase and Rational Engineering for Asymmetric Synthesis of beta-Amino Acids.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
3EB1
 
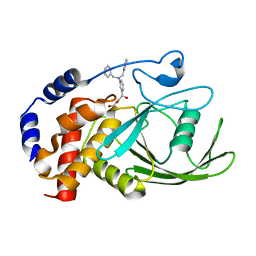 | | Crystal structure PTP1B complex with small molecule inhibitor LZP-25 | | Descriptor: | 4-[3-(dibenzylamino)phenyl]-2,4-dioxobutanoic acid, Tyrosine-protein phosphatase non-receptor type 1 | | Authors: | Liu, S, Zheng, L.-F, Wu, L, Yu, X, Xue, T, Gunawan, A.M, Long, Y.-Q, Zhang, Z.-Y. | | Deposit date: | 2008-08-26 | | Release date: | 2009-07-07 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Targeting inactive enzyme conformation: aryl diketoacid derivatives as a new class of PTP1B inhibitors.
J.Am.Chem.Soc., 130, 2008
|
|
5C8S
 
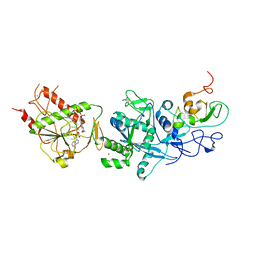 | | Crystal structure of the SARS coronavirus nsp14-nsp10 complex with functional ligands SAH and GpppA | | Descriptor: | GUANOSINE-P3-ADENOSINE-5',5'-TRIPHOSPHATE, Guanine-N7 methyltransferase, MAGNESIUM ION, ... | | Authors: | Ma, Y.Y, Wu, L.J, Zhang, R.G, Rao, Z.H. | | Deposit date: | 2015-06-26 | | Release date: | 2015-07-15 | | Last modified: | 2015-08-12 | | Method: | X-RAY DIFFRACTION (3.326 Å) | | Cite: | Structural basis and functional analysis of the SARS coronavirus nsp14-nsp10 complex
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
7PSK
 
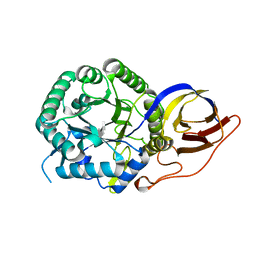 | | Crystal structure of beta-glucuronidase from Acidobacterium capsulatum in complex with covalent inhibitor GR109 | | Descriptor: | 2-acetamido-2-deoxy-6-O-sulfo-alpha-D-glucopyranose-(1-4)-(2R,3S,5R,6R)-2,3,4,5,6-pentakis(oxidanyl)cyclohexane-1-carboxylic acid, Beta-glucuronidase | | Authors: | Armstrong, Z, Wu, L, Davies, G.J. | | Deposit date: | 2021-09-23 | | Release date: | 2022-08-03 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.09 Å) | | Cite: | Mechanism-based heparanase inhibitors reduce cancer metastasis in vivo.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7PSI
 
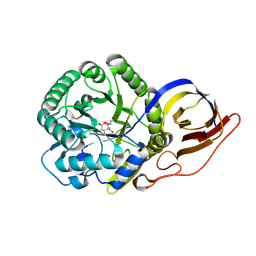 | | Crystal structure of beta-glucuronidase from Acidobacterium capsulatum in complex with covalent inhibitor ME727 | | Descriptor: | (2R,3S,5R,6R)-2,3,4,5,6-pentakis(oxidanyl)cyclohexane-1-carboxylic acid, Beta-glucuronidase, SULFATE ION | | Authors: | Armstrong, Z, Wu, L, Davies, G.J. | | Deposit date: | 2021-09-23 | | Release date: | 2022-08-03 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Mechanism-based heparanase inhibitors reduce cancer metastasis in vivo.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7PSH
 
 | |
7PSJ
 
 | | Crystal structure of beta-glucuronidase from Acidobacterium capsulatum in complex with covalent inhibitor VL166 | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose-(1-4)-(2R,3S,5R,6R)-2,3,4,5,6-pentakis(oxidanyl)cyclohexane-1-carboxylic acid, Beta-glucuronidase | | Authors: | Armstrong, Z, Wu, L, Davies, G.J. | | Deposit date: | 2021-09-23 | | Release date: | 2022-08-03 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Mechanism-based heparanase inhibitors reduce cancer metastasis in vivo.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
4GVJ
 
 | | Tyk2 (JH1) in complex with adenosine di-phosphate | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Non-receptor tyrosine-protein kinase TYK2 | | Authors: | Liang, J, Abbema, A.V, Bao, L, Barrett, K, Beresini, M, Berezhkovskiy, L, Blair, W, Chang, C, Driscoll, J, Eigenbrot, C, Ghilardi, N, Gibbons, P, Halladay, J, Johnson, A, Kohli, P.B, Lai, Y, Liimatta, M, Mantik, P, Menghrajani, K, Murray, J, Sambrone, A, Shao, Y, Shia, S, Shin, Y, Smith, J, Sohn, S, Stanley, M, Tsui, V, Ultsch, M, Wu, L, Zhang, B, Magnuson, S. | | Deposit date: | 2012-08-30 | | Release date: | 2013-08-14 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Lead identification of novel and selective TYK2 inhibitors.
Eur.J.Med.Chem., 67, 2013
|
|
7CZF
 
 | | Crystal structure of Kaposi Sarcoma associated herpesvirus (KSHV ) gHgL in complex with the ligand binding domian (LBD) of EphA2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Su, C, Wu, L.L, Song, H, Chai, Y, Qi, J.X, Yan, J.H, Gao, G.F. | | Deposit date: | 2020-09-08 | | Release date: | 2020-10-21 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Molecular basis of EphA2 recognition by gHgL from gammaherpesviruses.
Nat Commun, 11, 2020
|
|
7DL1
 
 | | The mutant E310G/G323S structure of 3,5-DAHDHcca complex with NADPH | | Descriptor: | 3,5-diaminohexanoate dehydrogenase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Liu, N, Wu, L, Zhu, D.M, Zhou, J.H. | | Deposit date: | 2020-11-25 | | Release date: | 2021-09-29 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.72 Å) | | Cite: | Crystal Structures and Catalytic Mechanism of l-erythro-3,5-Diaminohexanoate Dehydrogenase and Rational Engineering for Asymmetric Synthesis of beta-Amino Acids.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
5C8T
 
 | | Crystal structure of the SARS coronavirus nsp14-nsp10 complex with functional ligand SAM | | Descriptor: | Guanine-N7 methyltransferase, MAGNESIUM ION, Non-structural protein 10, ... | | Authors: | Ma, Y.Y, Wu, L.J, Zhang, R.G, Rao, Z.H. | | Deposit date: | 2015-06-26 | | Release date: | 2015-07-15 | | Last modified: | 2015-08-12 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural basis and functional analysis of the SARS coronavirus nsp14-nsp10 complex
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
5C8U
 
 | | Crystal structure of the SARS coronavirus nsp14-nsp10 complex | | Descriptor: | Guanine-N7 methyltransferase, MAGNESIUM ION, Non-structural protein 10, ... | | Authors: | Ma, Y.Y, Wu, L.J, Zhang, R.G, Rao, Z.H. | | Deposit date: | 2015-06-26 | | Release date: | 2015-07-15 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3.401 Å) | | Cite: | Structural basis and functional analysis of the SARS coronavirus nsp14-nsp10 complex
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
7DT1
 
 | | The structure of Lactobacillus fermentum 4,6-alpha-Glucanotransferase | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Yang, W.K, Yong, Y.H, Wu, L, Chen, S, Zhou, J.H, Wu, J. | | Deposit date: | 2021-01-04 | | Release date: | 2022-01-12 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.43002272 Å) | | Cite: | Characterization of a new 4,6-alpha-glucanotransferase from Limosilactobacillus fermentum NCC 3057 with ability of synthesizing low molecular mass isomalto-/maltopolysaccharide
Food Biosci, 46, 2022
|
|
5MU5
 
 | | Structure of MAf glycosyltransferase from Magnetospirillum magneticum AMB-1 | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, SULFATE ION, ... | | Authors: | Sulzenbacher, G, Roig-Zamboni, V, Murat, D, Vincentelli, R, Wu, L.F, Guerardel, Y, Alberto, F. | | Deposit date: | 2017-01-12 | | Release date: | 2017-11-15 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Glycosylate and move! The glycosyltransferase Maf is involved in bacterial flagella formation.
Environ. Microbiol., 20, 2018
|
|
8KFX
 
 | | Gi bound CCR8 complex with nonpeptide agonist LMD-009 | | Descriptor: | 8-[[3-(2-methoxyphenoxy)phenyl]methyl]-1-(2-phenylethyl)-1,3,8-triazaspiro[4.5]decan-4-one, C-C chemokine receptor type 8,Fusion protein, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Jiang, S, Lin, X, Wu, L.J, Xu, F. | | Deposit date: | 2023-08-16 | | Release date: | 2024-02-07 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (2.96 Å) | | Cite: | Unveiling the structural mechanisms of nonpeptide ligand recognition and activation in human chemokine receptor CCR8.
Sci Adv, 10, 2024
|
|
8KFZ
 
 | | Gi bound CCR8 in ligand free state | | Descriptor: | C-C chemokine receptor type 8,LgBiT fusion protein,Recombinant Human Rhinovirus, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Jiang, S, Lin, X, Wu, L.J, Xu, F. | | Deposit date: | 2023-08-16 | | Release date: | 2024-02-07 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Unveiling the structural mechanisms of nonpeptide ligand recognition and activation in human chemokine receptor CCR8.
Sci Adv, 10, 2024
|
|
