4JCM
 
 | | Crystal structure of Gamma-CGTASE from Alkalophilic bacillus clarkii at 1.65 Angstrom resolution | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Wu, L, Yang, D, Zhou, J, Wu, J, Chen, J. | | Deposit date: | 2013-02-22 | | Release date: | 2014-02-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | The Crystal Structure of Gamma-Cgtase from Alkalophilic Bacillus Clarkii at 1.65 Angstrom Resolution.
To be Published
|
|
7LZ4
 
 | | Crystal structure of A211D mutant of Protein Kinase A RIa subunit, a Carney Complex mutation | | Descriptor: | ADENOSINE-3',5'-CYCLIC-MONOPHOSPHATE, cAMP-dependent protein kinase type I-alpha regulatory subunit, N-terminally processed | | Authors: | Del Rio, J, Wu, J, Taylor, S.S. | | Deposit date: | 2021-03-08 | | Release date: | 2021-05-26 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (4.155 Å) | | Cite: | Noncanonical protein kinase A activation by oligomerization of regulatory subunits as revealed by inherited Carney complex mutations.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
1T2M
 
 | | Solution Structure Of The Pdz Domain Of AF-6 | | Descriptor: | AF-6 protein | | Authors: | Zhou, H, Wu, J.H, Xu, Y.Q, Huang, A.D, Shi, Y.Y. | | Deposit date: | 2004-04-22 | | Release date: | 2005-02-08 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of AF-6 PDZ Domain and Its Interaction with the C-terminal Peptides from Neurexin and Bcr
J.Biol.Chem., 280, 2005
|
|
2LNW
 
 | | Identification and structural basis for a novel interaction between Vav2 and Arap3 | | Descriptor: | Arf-GAP with Rho-GAP domain, ANK repeat and PH domain-containing protein 3, Guanine nucleotide exchange factor VAV2 | | Authors: | Wu, B, Zhang, J, Wu, J, Shi, Y. | | Deposit date: | 2012-01-05 | | Release date: | 2012-11-21 | | Last modified: | 2024-10-16 | | Method: | SOLUTION NMR | | Cite: | Identification and structural basis for a novel interaction between Vav2 and Arap3.
J.Struct.Biol., 180, 2012
|
|
2LNX
 
 | | Solution structure of Vav2 SH2 domain | | Descriptor: | Guanine nucleotide exchange factor VAV2 | | Authors: | Wu, B, Zhang, J, Wu, J, Shi, Y. | | Deposit date: | 2012-01-05 | | Release date: | 2012-11-21 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Identification and structural basis for a novel interaction between Vav2 and Arap3.
J.Struct.Biol., 180, 2012
|
|
4YHC
 
 | | Crystal structure of the WD40 domain of SCAP from fission yeast | | Descriptor: | CITRIC ACID, Sterol regulatory element-binding protein cleavage-activating protein | | Authors: | Gong, X, Li, J.X, Wu, J.P, Yan, C.Y, Yan, N. | | Deposit date: | 2015-02-27 | | Release date: | 2015-04-01 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structure of the WD40 domain of SCAP from fission yeast reveals the molecular basis for SREBP recognition.
Cell Res., 25, 2015
|
|
4O26
 
 | |
6VGU
 
 | |
6NE7
 
 | | Structure of G810A mutant of RET protein tyrosine kinase domain. | | Descriptor: | ADENOSINE MONOPHOSPHATE, FORMIC ACID, Proto-oncogene tyrosine-protein kinase receptor Ret | | Authors: | Terzyan, S.S, Shen, T, Wu, J, Mooers, B.H.M. | | Deposit date: | 2018-12-17 | | Release date: | 2019-06-05 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Structural basis of resistance of mutant RET protein-tyrosine kinase to its inhibitors nintedanib and vandetanib.
J.Biol.Chem., 294, 2019
|
|
6NO7
 
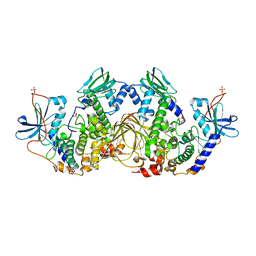 | | Crystal Structure of the full-length wild-type PKA RIa Holoenzyme | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, cAMP-dependent protein kinase catalytic subunit alpha, ... | | Authors: | Lu, T, Wu, J, Taylor, S.S. | | Deposit date: | 2019-01-15 | | Release date: | 2019-07-24 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.55 Å) | | Cite: | Two PKA RI alpha holoenzyme states define ATP as an isoform-specific orthosteric inhibitor that competes with the allosteric activator, cAMP.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
1OZJ
 
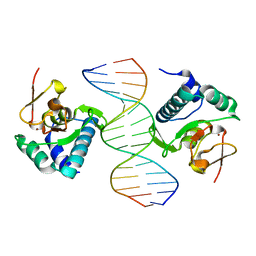 | | Crystal structure of Smad3-MH1 bound to DNA at 2.4 A resolution | | Descriptor: | SMAD 3, Smad binding element, ZINC ION | | Authors: | Chai, J, Wu, J.-W, Yan, N, Massague, J, Pavletich, N.P, Shi, Y. | | Deposit date: | 2003-04-09 | | Release date: | 2004-03-23 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Features of a Smad3 MH1-DNA complex. Roles of water and zinc in DNA binding.
J.Biol.Chem., 278, 2003
|
|
6NEC
 
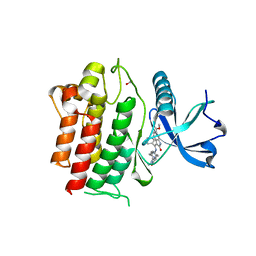 | | STRUCTURE OF RET PROTEIN TYROSINE KINASE DOMAIN IN COMPLEX WITH NINTEDANIB | | Descriptor: | FORMIC ACID, Proto-oncogene tyrosine-protein kinase receptor Ret, methyl (3Z)-3-{[(4-{methyl[(4-methylpiperazin-1-yl)acetyl]amino}phenyl)amino](phenyl)methylidene}-2-oxo-2,3-dihydro-1H-indole-6-carboxylate | | Authors: | Terzyan, S.S, Shen, T, Wu, J, Mooers, B.H.M. | | Deposit date: | 2018-12-17 | | Release date: | 2019-06-05 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Structural basis of resistance of mutant RET protein-tyrosine kinase to its inhibitors nintedanib and vandetanib.
J.Biol.Chem., 294, 2019
|
|
6NJA
 
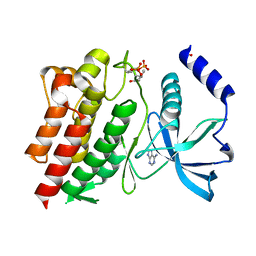 | | Structure of WT RET protein tyrosine kinase domain at 1.92A resolution. | | Descriptor: | ADENINE, FORMIC ACID, Proto-oncogene tyrosine-protein kinase receptor Ret | | Authors: | Terzyan, S.S, Shen, T, Wu, J, Mooers, B.H.M. | | Deposit date: | 2019-01-02 | | Release date: | 2019-06-05 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Structural basis of resistance of mutant RET protein-tyrosine kinase to its inhibitors nintedanib and vandetanib.
J.Biol.Chem., 294, 2019
|
|
6OJN
 
 | | Comparative Model of SGIV Major Coat Protein (MCP) Trimer Based on Cryo-EM Map | | Descriptor: | Major capsid protein | | Authors: | Pintilie, G, Chen, D.-H, Tran, B.N, Jakana, J, Wu, J, Hew, C.L, Chiu, W. | | Deposit date: | 2019-04-11 | | Release date: | 2019-06-12 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (8.6 Å) | | Cite: | Segmentation and Comparative Modeling in an 8.6- angstrom Cryo-EM Map of the Singapore Grouper Iridovirus.
Structure, 27, 2019
|
|
3JCO
 
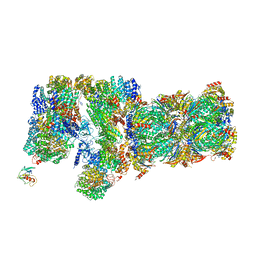 | | Structure of yeast 26S proteasome in M1 state derived from Titan dataset | | Descriptor: | 26S protease regulatory subunit 4 homolog, 26S protease regulatory subunit 6A, 26S protease regulatory subunit 6B homolog, ... | | Authors: | Luan, B, Huang, X.L, Wu, J.P, Shi, Y.G, Wang, F. | | Deposit date: | 2016-01-06 | | Release date: | 2016-06-15 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (4.8 Å) | | Cite: | Structure of an endogenous yeast 26S proteasome reveals two major conformational states.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
4DSO
 
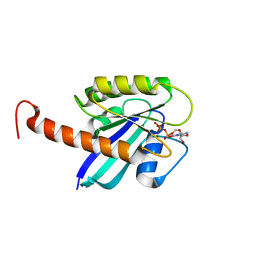 | | Small-molecule ligands bind to a distinct pocket in Ras and inhibit SOS-mediated nucleotide exchange activity | | Descriptor: | 5'-GUANOSINE-DIPHOSPHATE-MONOTHIOPHOSPHATE, BENZAMIDINE, GLYCEROL, ... | | Authors: | Oh, A, Maurer, T, Garrenton, L.S, Pitts, K, Anderson, D.J, Skelton, N.J, Fauber, B.P, Pan, B, Malek, S, Stokoe, D, Ludlam, M, Bowman, K.K, Wu, J, Giannetti, A.M, Starovasnik, M.A, Mellman, I, Jackson, P.K, Ruldolph, J, Fang, G, Wang, W. | | Deposit date: | 2012-02-19 | | Release date: | 2012-04-04 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Small-molecule ligands bind to a distinct pocket in Ras and inhibit SOS-mediated nucleotide exchange activity.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4DSU
 
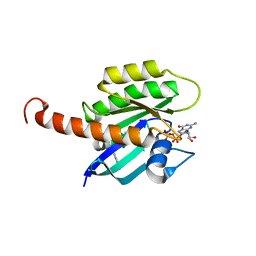 | | Small-molecule ligands bind to a distinct pocket in Ras and inhibit SOS-mediated nucleotide exchange activity | | Descriptor: | BENZIMIDAZOLE, GTPase KRas, isoform 2B, ... | | Authors: | Oh, A, Maurer, T, Garrenton, L.S, Pitts, K, Anderson, D.J, Skelton, N.J, Fauber, B.P, Pan, B, Malek, S, Stokoe, D, Ludlam, M, Bowman, K.K, Wu, J, Giannetti, A.M, Starovasnik, M.A, Mellman, I, Jackson, P.K, Ruldolph, J, Fang, G, Wang, W. | | Deposit date: | 2012-02-19 | | Release date: | 2012-04-04 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Small-molecule ligands bind to a distinct pocket in Ras and inhibit SOS-mediated nucleotide exchange activity.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4DST
 
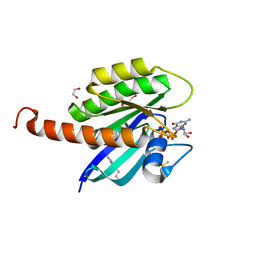 | | Small-molecule ligands bind to a distinct pocket in Ras and inhibit SOS-mediated nucleotide exchange activity | | Descriptor: | 1,2-ETHANEDIOL, 2-(4,6-dichloro-2-methyl-1H-indol-3-yl)ethanamine, ACETATE ION, ... | | Authors: | Oh, A, Maurer, T, Garrenton, L.S, Pitts, K, Anderson, D.J, Skelton, N.J, Fauber, B.P, Pan, B, Malek, S, Stokoe, D, Ludlam, M, Bowman, K.K, Wu, J, Giannetti, A.M, Starovasnik, M.A, Mellman, I, Jackson, P.K, Ruldolph, J, Fang, G, Wang, W. | | Deposit date: | 2012-02-19 | | Release date: | 2012-04-04 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Small-molecule ligands bind to a distinct pocket in Ras and inhibit SOS-mediated nucleotide exchange activity.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4DSN
 
 | | Small-molecule ligands bind to a distinct pocket in Ras and inhibit SOS-mediated nucleotide exchange activity | | Descriptor: | 1,2-ETHANEDIOL, GTPase KRas, isoform 2B, ... | | Authors: | Oh, A, Maurer, T, Garrenton, L.S, Pitts, K, Anderson, D.J, Skelton, N.J, Fauber, B.P, Pan, B, Malek, S, Stokoe, D, Ludlam, M, Bowman, K.K, Wu, J, Giannetti, A.M, Starovasnik, M.A, Mellman, I, Jackson, P.K, Ruldolph, J, Fang, G, Wang, W. | | Deposit date: | 2012-02-19 | | Release date: | 2012-04-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Small-molecule ligands bind to a distinct pocket in Ras and inhibit SOS-mediated nucleotide exchange activity.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
7Y11
 
 | | Crystal structure of AtSFH5-Sec14 in complex with egg PA | | Descriptor: | (2R)-1-(hexadecanoyloxy)-3-(phosphonooxy)propan-2-yl (9Z)-octadec-9-enoate, CHLORIDE ION, NICKEL (II) ION, ... | | Authors: | Lu, Y.Q, Wang, X.Q, Luo, Z.P, Wu, J.W. | | Deposit date: | 2022-06-06 | | Release date: | 2023-04-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Arabidopsis Sec14 proteins (SFH5 and SFH7) mediate interorganelle transport of phosphatidic acid and regulate chloroplast development.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
7Y10
 
 | | Crystal structure of AtSFH5-Sec14 in complex with DPPA | | Descriptor: | 1,2-DIPALMITOYL-SN-GLYCERO-3-PHOSPHATE, NICKEL (II) ION, Phosphatidylinositol/phosphatidylcholine transfer protein SFH5 | | Authors: | Lu, Y.Q, Wang, X.Q, Luo, Z.P, Wu, J.W. | | Deposit date: | 2022-06-06 | | Release date: | 2023-05-31 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Arabidopsis Sec14 proteins (SFH5 and SFH7) mediate interorganelle transport of phosphatidic acid and regulate chloroplast development.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
2LKZ
 
 | | Solution structure of the second RRM domain of RBM5 | | Descriptor: | RNA-binding protein 5 | | Authors: | Song, Z, Wu, P, Zhang, J, Wu, J, Shi, Y. | | Deposit date: | 2011-10-23 | | Release date: | 2012-08-08 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the second RRM domain of RBM5 and its unusual binding characters for different RNA targets
Biochemistry, 2012
|
|
1J3H
 
 | | Crystal structure of apoenzyme cAMP-dependent protein kinase catalytic subunit | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, cAMP-dependent protein kinase, alpha-catalytic subunit | | Authors: | Akamine, P, Madhusudan, Wu, J, Xuong, N.H, Ten Eyck, L.F, Taylor, S.S. | | Deposit date: | 2003-01-31 | | Release date: | 2003-03-04 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Dynamic Features of cAMP-dependent Protein Kinase Revealed by Apoenzyme Crystal Structure
J.Mol.Biol., 327, 2003
|
|
2ZN7
 
 | | CRYSTAL STRUCTURES OF PTP1B-Inhibitor Complexes | | Descriptor: | 4-bromo-3-(carboxymethoxy)-5-{3-[cyclohexyl(phenylcarbonyl)amino]phenyl}thiophene-2-carboxylic acid, Tyrosine-protein phosphatase non-receptor type 1 | | Authors: | Xu, W, Wu, J. | | Deposit date: | 2008-04-22 | | Release date: | 2008-10-07 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure-based optimization of protein tyrosine phosphatase-1 B inhibitors: capturing interactions with arginine 24
Chemmedchem, 3, 2008
|
|
1A8K
 
 | | CRYSTALLOGRAPHIC ANALYSIS OF HUMAN IMMUNODEFICIENCY VIRUS 1 PROTEASE WITH AN ANALOG OF THE CONSERVED CA-P2 SUBSTRATE: INTERACTIONS WITH FREQUENTLY OCCURRING GLUTAMIC ACID RESIDUE AT P2' POSITION OF SUBSTRATES | | Descriptor: | HIV PROTEASE, N-[(2R)-2-({N~5~-[amino(iminio)methyl]-L-ornithyl-L-valyl}amino)-4-methylpentyl]-L-phenylalanyl-L-alpha-glutamyl-L-alanyl-L-norleucinamide | | Authors: | Weber, I.T, Wu, J, Adomat, J, Harrison, R.W, Kimmel, A.R, Wondrak, E.M, Louis, J.M. | | Deposit date: | 1998-03-27 | | Release date: | 1999-01-13 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystallographic analysis of human immunodeficiency virus 1 protease with an analog of the conserved CA-p2 substrate -- interactions with frequently occurring glutamic acid residue at P2' position of substrates.
Eur.J.Biochem., 249, 1997
|
|
