5YWA
 
 | |
7E1L
 
 | | Crystal structure of apo form PhlH | | Descriptor: | DUF1956 domain-containing protein | | Authors: | Zhang, N, Wu, J, He, Y.X, Ge, H. | | Deposit date: | 2021-02-01 | | Release date: | 2022-02-02 | | Last modified: | 2022-08-24 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Molecular basis for coordinating secondary metabolite production by bacterial and plant signaling molecules.
J.Biol.Chem., 298, 2022
|
|
5X6G
 
 | | Crystal Structure of SMAD5-MH1/palindromic SBE DNA complex | | Descriptor: | DNA (5'-D(P*AP*TP*CP*AP*GP*TP*CP*TP*AP*GP*AP*CP*AP*TP*A)-3'), DNA (5'-D(P*GP*TP*AP*TP*GP*TP*CP*TP*AP*GP*AP*CP*TP*GP*A)-3'), Mothers against decapentaplegic homolog 5, ... | | Authors: | Chai, N, Wang, J, Wang, Z.X, Wu, J.W. | | Deposit date: | 2017-02-21 | | Release date: | 2017-03-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Structural basis for the Smad5 MH1 domain to recognize different DNA sequences.
Nucleic Acids Res., 43, 2015
|
|
5X6H
 
 | | Crystal Structure of SMAD5-MH1/GC-BRE DNA complex | | Descriptor: | DNA (5'-D(P*GP*TP*AP*TP*GP*GP*CP*GP*CP*CP*AP*TP*AP*C)-3'), Mothers against decapentaplegic homolog 5, ZINC ION | | Authors: | Chai, N, Wang, J, Wang, Z.X, Wu, J.W. | | Deposit date: | 2017-02-22 | | Release date: | 2017-03-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural basis for the Smad5 MH1 domain to recognize different DNA sequences.
Nucleic Acids Res., 43, 2015
|
|
7D65
 
 | | Cryo-EM Structure of human CALHM5 in the presence of Ca2+ | | Descriptor: | 1,2-DIOCTANOYL-SN-GLYCERO-3-PHOSPHATE, Calcium homeostasis modulator protein 5 | | Authors: | Liu, J, Guan, F.H, Wu, J, Wan, F.T, Lei, M, Ye, S. | | Deposit date: | 2020-09-29 | | Release date: | 2020-12-23 | | Method: | ELECTRON MICROSCOPY (2.94 Å) | | Cite: | Cryo-EM structures of human calcium homeostasis modulator 5.
Cell Discov, 6, 2020
|
|
7D61
 
 | | Cryo-EM Structure of human CALHM5 in the presence of EDTA | | Descriptor: | 1,2-DIOCTANOYL-SN-GLYCERO-3-PHOSPHATE, Calcium homeostasis modulator protein 5 | | Authors: | Liu, J, Guan, F.H, Wu, J, Wan, F.T, Lei, M, Ye, S. | | Deposit date: | 2020-09-28 | | Release date: | 2020-12-23 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Cryo-EM structures of human calcium homeostasis modulator 5.
Cell Discov, 6, 2020
|
|
5WZK
 
 | | Structure of APUM23-deletion-of-insert-region-GGAAUUGACGG | | Descriptor: | Pumilio homolog 23, RNA (5'-R(*GP*GP*AP*AP*UP*UP*GP*AP*CP*GP*G)-3') | | Authors: | Bao, H, Wang, N, Wang, C, Jiang, Y, Wu, J, Shi, Y. | | Deposit date: | 2017-01-18 | | Release date: | 2017-09-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural basis for the specific recognition of 18S rRNA by APUM23.
Nucleic Acids Res., 45, 2017
|
|
5SYO
 
 | | Crystal structure of a chimeric acetylcholine binding protein from Aplysia californica (Ac-AChBP) containing loop C from the human alpha 3 nicotinic acetylcholine receptor in complex with Cytisine | | Descriptor: | (1R,5S)-1,2,3,4,5,6-HEXAHYDRO-8H-1,5-METHANOPYRIDO[1,2-A][1,5]DIAZOCIN-8-ONE, Soluble acetylcholine receptor, Neuronal acetylcholine receptor subunit alpha-3 chimera | | Authors: | Bobango, J, Wu, J, Talley, I.T, Talley, T.T. | | Deposit date: | 2016-08-11 | | Release date: | 2016-10-12 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of a chimeric acetylcholine binding protein from Aplysia californica (Ac-AChBP) containing loop C from the human alpha 3 nicotinic acetylcholine receptor in complex with Cytisine
To Be Published
|
|
7C7A
 
 | | Cryo-EM structure of yeast Ribonuclease MRP with substrate ITS1 | | Descriptor: | Internal Transcribed Spacer 1, MAGNESIUM ION, RNases MRP/P 32.9 kDa subunit, ... | | Authors: | Lan, P, Wu, J, Lei, M. | | Deposit date: | 2020-05-24 | | Release date: | 2020-07-08 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structural insight into precursor ribosomal RNA processing by ribonuclease MRP.
Science, 369, 2020
|
|
4WV9
 
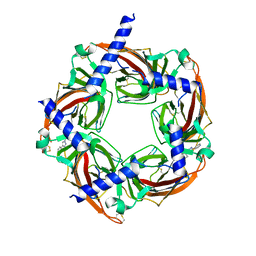 | | Crystal structure of acetylcholine binding protein (AChBP) from Aplysia Californica in complex with click chemistry compound (3-exo)-8,8-dimethyl-3-[4-(pyridin-4-yl)-1H-1,2,3-triazol-1-yl]-8-azoniabicyclo[3.2.1]octane | | Descriptor: | (3-exo)-8,8-dimethyl-3-[4-(pyridin-4-yl)-1H-1,2,3-triazol-1-yl]-8-azoniabicyclo[3.2.1]octane, Soluble acetylcholine receptor | | Authors: | Talley, T.T, Bobango, J, Wu, J.M, Sankaran, B. | | Deposit date: | 2014-11-04 | | Release date: | 2015-04-22 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of acetylcholine binding protein (AChBP) from Aplysia Californica in complex with click chemistry compound.
To Be Published
|
|
5YYZ
 
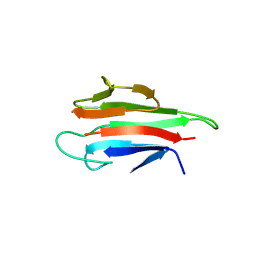 | | Crystal structure of the MEK1 FHA domain in complex with the HOP1 pThr318 peptide. | | Descriptor: | Meiosis-specific protein HOP1, Meiosis-specific serine/threonine-protein kinase MEK1 | | Authors: | Xie, C, Li, F, Jiang, Y, Wu, J, Shi, Y. | | Deposit date: | 2017-12-11 | | Release date: | 2018-10-17 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.798 Å) | | Cite: | Structural insights into the recognition of phosphorylated Hop1 by Mek1
Acta Crystallogr D Struct Biol, 74, 2018
|
|
5Z1F
 
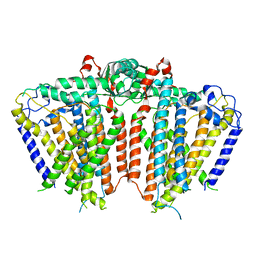 | |
5YWD
 
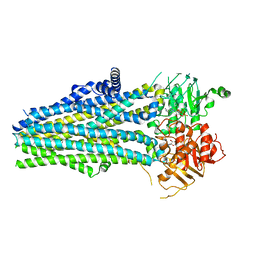 | |
7D3E
 
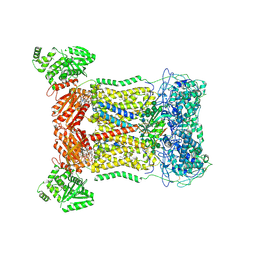 | | Cryo-EM structure of human DUOX1-DUOXA1 in low-calcium state | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Dual oxidase 1, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Chen, L, Wu, J.X. | | Deposit date: | 2020-09-19 | | Release date: | 2020-12-09 | | Last modified: | 2021-06-23 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structures of human dual oxidase 1 complex in low-calcium and high-calcium states.
Nat Commun, 12, 2021
|
|
5YW7
 
 | |
7D3F
 
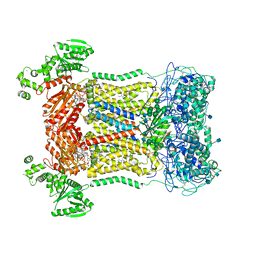 | | Cryo-EM structure of human DUOX1-DUOXA1 in high-calcium state | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Dual oxidase 1, ... | | Authors: | Chen, L, Wu, J.X. | | Deposit date: | 2020-09-19 | | Release date: | 2020-12-09 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Structures of human dual oxidase 1 complex in low-calcium and high-calcium states.
Nat Commun, 12, 2021
|
|
5YYX
 
 | | Crystal Structure of the MEK1 FHA domain | | Descriptor: | Meiosis-specific serine/threonine-protein kinase MEK1 | | Authors: | Xie, C, Li, F, Jiang, Y, Wu, J, Shi, Y. | | Deposit date: | 2017-12-11 | | Release date: | 2018-10-10 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.684 Å) | | Cite: | Structural insights into the recognition of phosphorylated Hop1 by Mek1
Acta Crystallogr D Struct Biol, 74, 2018
|
|
7C79
 
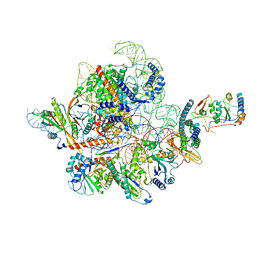 | | Cryo-EM structure of yeast Ribonuclease MRP | | Descriptor: | MAGNESIUM ION, RNases MRP/P 32.9 kDa subunit, Ribonuclease MRP RNA subunit NME1, ... | | Authors: | Lan, P, Wu, J, Lei, M. | | Deposit date: | 2020-05-24 | | Release date: | 2020-07-08 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Structural insight into precursor ribosomal RNA processing by ribonuclease MRP.
Science, 369, 2020
|
|
5T0I
 
 | | Structural basis for dynamic regulation of the human 26S proteasome | | Descriptor: | 26S protease regulatory subunit 10B, 26S protease regulatory subunit 4, 26S protease regulatory subunit 6A, ... | | Authors: | Chen, S, Wu, J, Lu, Y, Ma, Y.B, Lee, B.H, Yu, Z, Ouyang, Q, Finley, D, Kirschner, M.W, Mao, Y. | | Deposit date: | 2016-08-16 | | Release date: | 2016-10-19 | | Last modified: | 2016-11-30 | | Method: | ELECTRON MICROSCOPY (8 Å) | | Cite: | Structural basis for dynamic regulation of the human 26S proteasome.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5T0C
 
 | | Structural basis for dynamic regulation of the human 26S proteasome | | Descriptor: | 26S protease regulatory subunit 10B, 26S protease regulatory subunit 4, 26S protease regulatory subunit 6A, ... | | Authors: | Chen, S, Wu, J, Lu, Y, Ma, Y.B, Lee, B.H, Yu, Z, Ouyang, Q, Finley, D, Kirschner, M.W, Mao, Y. | | Deposit date: | 2016-08-15 | | Release date: | 2016-10-19 | | Last modified: | 2018-07-18 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structural basis for dynamic regulation of the human 26S proteasome.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5T0J
 
 | | Structural basis for dynamic regulation of the human 26S proteasome | | Descriptor: | 26S protease regulatory subunit 10B, 26S protease regulatory subunit 4, 26S protease regulatory subunit 6A, ... | | Authors: | Chen, S, Wu, J, Lu, Y, Ma, Y.B, Lee, B.H, Yu, Z, Ouyang, Q, Finley, D, Kirschner, M.W, Mao, Y. | | Deposit date: | 2016-08-16 | | Release date: | 2016-10-19 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (8 Å) | | Cite: | Structural basis for dynamic regulation of the human 26S proteasome.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5T0G
 
 | | Structural basis for dynamic regulation of the human 26S proteasome | | Descriptor: | 26S protease regulatory subunit 10B, 26S protease regulatory subunit 4, 26S protease regulatory subunit 6A, ... | | Authors: | Chen, S, Wu, J, Lu, Y, Ma, Y.B, Lee, B.H, Yu, Z, Ouyang, Q, Finley, D, Kirschner, M.W, Mao, Y. | | Deposit date: | 2016-08-16 | | Release date: | 2016-10-19 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Structural basis for dynamic regulation of the human 26S proteasome.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5T0H
 
 | | Structural basis for dynamic regulation of the human 26S proteasome | | Descriptor: | 26S protease regulatory subunit 10B, 26S protease regulatory subunit 4, 26S protease regulatory subunit 6A, ... | | Authors: | Chen, S, Wu, J, Lu, Y, Ma, Y.B, Lee, B.H, Yu, Z, Ouyang, Q, Finley, D, Kirschner, M.W, Mao, Y. | | Deposit date: | 2016-08-16 | | Release date: | 2016-10-19 | | Last modified: | 2016-11-30 | | Method: | ELECTRON MICROSCOPY (6.8 Å) | | Cite: | Structural basis for dynamic regulation of the human 26S proteasome.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
4ZJS
 
 | | Crystal structure of a chimeric acetylcholine binding protein from Aplysia Californica (Ac-AChBP) containing the main immunogenic region (MIR) from the human alpha 1 subunit of the muscle nicotinic acetylcholine receptor in complex with anatoxin-A. | | Descriptor: | 1-[(1R,6R)-9-azabicyclo[4.2.1]non-2-en-2-yl]ethanone, Acetylcholine receptor subunit alpha,Soluble acetylcholine receptor,Acetylcholine receptor subunit alpha,Soluble acetylcholine receptor | | Authors: | Talley, T.T, Bobango, J, Wu, J, Park, J.F, Luo, J, Lindsatrom, J, Taylor, P. | | Deposit date: | 2015-04-29 | | Release date: | 2015-05-13 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.2301 Å) | | Cite: | Main immunogenic region structure promotes binding of conformation-dependent myasthenia gravis autoantibodies, nicotinic acetylcholine receptor conformation maturation, and agonist sensitivity.
J. Neurosci., 29, 2009
|
|
4ZRU
 
 | | X-ray crystal structure of Lymnaea stagnalis acetylcholine binding protein (Ls-AChBP) in complex with 3-[2-[(2S)-pyrrolidin-2-yl]ethynyl]pyridine (TI-5180) | | Descriptor: | 3-[(2S)-pyrrolidin-2-ylethynyl]pyridine, Acetylcholine-binding protein, PHOSPHATE ION | | Authors: | Bobango, J, Sankaran, B, Park, J.F, Wu, J, Talley, T.T. | | Deposit date: | 2015-05-12 | | Release date: | 2015-05-27 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Comparisons of Binding Affinities for Neuronal Nicotinic Receptors (NNRs) and AChBPs, and Structural Features of a High-Affinity, Non-selective NNR Ligand-AChBP Co-crystal Structure
To be Published
|
|
