4P4H
 
 | |
3OE9
 
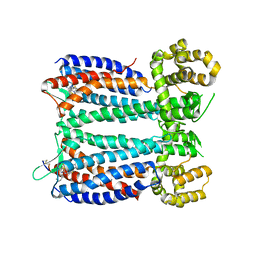 | | Crystal structure of the chemokine CXCR4 receptor in complex with a small molecule antagonist IT1t in P1 spacegroup | | Descriptor: | (6,6-dimethyl-5,6-dihydroimidazo[2,1-b][1,3]thiazol-3-yl)methyl N,N'-dicyclohexylimidothiocarbamate, C-X-C chemokine receptor type 4, Lysozyme Chimera | | Authors: | Wu, B, Mol, C.D, Han, G.W, Katritch, V, Chien, E.Y.T, Liu, W, Cherezov, V, Stevens, R.C, Accelerated Technologies Center for Gene to 3D Structure (ATCG3D), GPCR Network (GPCR) | | Deposit date: | 2010-08-12 | | Release date: | 2010-10-27 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structures of the CXCR4 chemokine GPCR with small-molecule and cyclic peptide antagonists.
Science, 330, 2010
|
|
4GL2
 
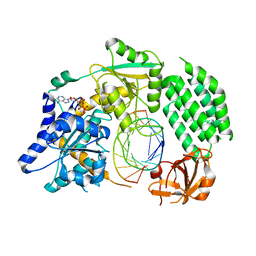 | | Structural Basis for dsRNA duplex backbone recognition by MDA5 | | Descriptor: | Interferon-induced helicase C domain-containing protein 1, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, RNA (5'-R(*AP*UP*CP*CP*GP*CP*GP*GP*CP*CP*CP*U)-3'), ... | | Authors: | Wu, B, Hur, S. | | Deposit date: | 2012-08-13 | | Release date: | 2013-01-09 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.557 Å) | | Cite: | Structural Basis for dsRNA Recognition, Filament Formation, and Antiviral Signal Activation by MDA5.
Cell(Cambridge,Mass.), 152, 2013
|
|
7W0I
 
 | |
3OE8
 
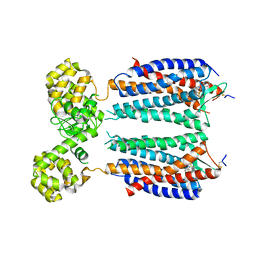 | | Crystal structure of the CXCR4 chemokine receptor in complex with a small molecule antagonist IT1t in P1 spacegroup | | Descriptor: | (6,6-dimethyl-5,6-dihydroimidazo[2,1-b][1,3]thiazol-3-yl)methyl N,N'-dicyclohexylimidothiocarbamate, C-X-C chemokine receptor type 4, Lysozyme Chimera | | Authors: | Wu, B, Mol, C.D, Han, G.W, Katritch, V, Chien, E.Y.T, Liu, W, Cherezov, V, Stevens, R.C, Accelerated Technologies Center for Gene to 3D Structure (ATCG3D), GPCR Network (GPCR) | | Deposit date: | 2010-08-12 | | Release date: | 2010-10-27 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structures of the CXCR4 chemokine GPCR with small-molecule and cyclic peptide antagonists.
Science, 330, 2010
|
|
5YRN
 
 | | Structure of RIP2 CARD domain | | Descriptor: | Receptor-interacting serine/threonine-protein kinase 2 | | Authors: | Wu, B, Gong, Q. | | Deposit date: | 2017-11-09 | | Release date: | 2018-11-14 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Structural basis of RIP2 activation and signaling.
Nat Commun, 9, 2018
|
|
5ZUU
 
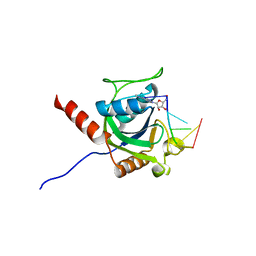 | | Crystal structure of AtCPSF30 YTH domain in complex with 10mer m6A-modified RNA | | Descriptor: | 30-kDa cleavage and polyadenylation specificity factor 30, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, GLYCEROL, ... | | Authors: | Wu, B, Nie, H, Li, S, Patel, D.J. | | Deposit date: | 2018-05-08 | | Release date: | 2019-05-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | CPSF30-L-mediated recognition of mRNA m6A modification controls alternative polyadenylation of nitrate signaling-related gene transcripts in Arabidopsis.
Mol Plant, 2021
|
|
5Z28
 
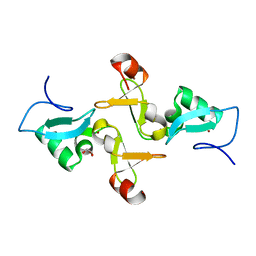 | | AtVAL2 PHD-Like domain | | Descriptor: | B3 domain-containing transcription repressor VAL2, GLYCEROL, ZINC ION | | Authors: | Wu, B.X, Zhang, M.M. | | Deposit date: | 2017-12-30 | | Release date: | 2018-05-02 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.245 Å) | | Cite: | Crystal structure of AtVAL12 PHD-Like domain
To Be Published
|
|
3ODU
 
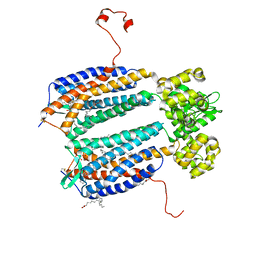 | | The 2.5 A structure of the CXCR4 chemokine receptor in complex with small molecule antagonist IT1t | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (6,6-dimethyl-5,6-dihydroimidazo[2,1-b][1,3]thiazol-3-yl)methyl N,N'-dicyclohexylimidothiocarbamate, C-X-C chemokine receptor type 4, ... | | Authors: | Wu, B, Mol, C.D, Han, G.W, Katritch, V, Chien, E.Y.T, Liu, W, Cherezov, V, Stevens, R.C, Accelerated Technologies Center for Gene to 3D Structure (ATCG3D), GPCR Network (GPCR) | | Deposit date: | 2010-08-11 | | Release date: | 2010-10-27 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structures of the CXCR4 chemokine GPCR with small-molecule and cyclic peptide antagonists.
Science, 330, 2010
|
|
7DUS
 
 | |
5YZY
 
 | | AtVAL1 B3 domain in complex with 13bp-DNA | | Descriptor: | B3 domain-containing transcription repressor VAL1, DNA (5'-D(*AP*TP*TP*CP*TP*GP*CP*AP*TP*GP*GP*AP*T)-3'), DNA (5'-D(*TP*AP*TP*CP*CP*AP*TP*GP*CP*AP*GP*AP*A)-3'), ... | | Authors: | Wu, B.X, Zhang, M.M. | | Deposit date: | 2017-12-17 | | Release date: | 2018-05-02 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Structural insight into the role of VAL1 B3 domain for targeting to FLC locus in Arabidopsis thaliana.
Biochem. Biophys. Res. Commun., 2018
|
|
5Z00
 
 | | AtVAL1 B3 domain in complex with 15bp-DNA | | Descriptor: | B3 domain-containing transcription repressor VAL1, DNA (5'-D(*AP*AP*TP*TP*CP*TP*GP*CP*AP*TP*GP*GP*AP*TP*T)-3'), DNA (5'-D(*TP*AP*AP*TP*CP*CP*AP*TP*GP*CP*AP*GP*AP*AP*T)-3'), ... | | Authors: | Wu, B.X, Zhang, M.M. | | Deposit date: | 2017-12-17 | | Release date: | 2018-05-02 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.587 Å) | | Cite: | Structural insight into the role of VAL1 B3 domain for targeting to FLC locus in Arabidopsis thaliana.
Biochem. Biophys. Res. Commun., 2018
|
|
5YZZ
 
 | | AtVAL1 B3 domain in complex with 13bp-DNA | | Descriptor: | B3 domain-containing transcription repressor VAL1, DNA (5'-D(*AP*TP*TP*CP*TP*GP*CP*AP*TP*GP*GP*AP*T)-3'), DNA (5'-D(*TP*AP*TP*CP*CP*AP*TP*GP*CP*AP*GP*AP*A)-3') | | Authors: | Wu, B.X, Zhang, M.M. | | Deposit date: | 2017-12-17 | | Release date: | 2018-05-02 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.58 Å) | | Cite: | Structural insight into the role of VAL1 B3 domain for targeting to FLC locus in Arabidopsis thaliana.
Biochem. Biophys. Res. Commun., 2018
|
|
8KA3
 
 | |
8KA5
 
 | |
8YHK
 
 | | The Crystal Structure of P21-Activated Kinases Pak4 from Biortus | | Descriptor: | Serine/threonine-protein kinase PAK 4, TRIETHYLENE GLYCOL | | Authors: | Wang, F, Cheng, W, Lv, Z, Qi, J, Wu, B. | | Deposit date: | 2024-02-28 | | Release date: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | The Crystal Structure of P21-Activated Kinases Pak4 from Biortus
To Be Published
|
|
4MBS
 
 | | Crystal Structure of the CCR5 Chemokine Receptor | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 4,4-difluoro-N-[(1S)-3-{(3-exo)-3-[3-methyl-5-(propan-2-yl)-4H-1,2,4-triazol-4-yl]-8-azabicyclo[3.2.1]oct-8-yl}-1-phenylpropyl]cyclohexanecarboxamide, Chimera protein of C-C chemokine receptor type 5 and Rubredoxin, ... | | Authors: | Tan, Q, Zhu, Y, Han, G.W, Li, J, Fenalti, G, Liu, H, Cherezov, V, Stevens, R.C, GPCR Network (GPCR), Zhao, Q, Wu, B. | | Deposit date: | 2013-08-19 | | Release date: | 2013-09-11 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | Structure of the CCR5 chemokine receptor-HIV entry inhibitor maraviroc complex.
Science, 341, 2013
|
|
8JRU
 
 | | Cryo-EM structure of the glucagon receptor bound to beta-arrestin 1 in ligand-free state | | Descriptor: | Beta-arrestin 1 and single-chain fragment variable 30 (scFv30), HA signal peptide,HPC4 purification tag,Glucagon receptor,C-terminal tail of Vasopressin V2 receptor, Nanobody 32, ... | | Authors: | Chen, K, Zhang, C, Lin, S, Zhao, Q, Wu, B. | | Deposit date: | 2023-06-17 | | Release date: | 2023-08-16 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Tail engagement of arrestin at the glucagon receptor.
Nature, 620, 2023
|
|
8JRV
 
 | | Cryo-EM structure of the glucagon receptor bound to glucagon and beta-arrestin 1 | | Descriptor: | Beta-arrestin 1 and single-chain fragment variable 30 (scFv30), Glucagon, HA signal peptide,HPC4 purification tag,Glucagon receptor,C-terminal tail of Vasopressin V2 receptor, ... | | Authors: | Chen, K, Zhang, C, Lin, S, Zhao, Q, Wu, B. | | Deposit date: | 2023-06-17 | | Release date: | 2023-08-16 | | Last modified: | 2023-09-13 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Tail engagement of arrestin at the glucagon receptor.
Nature, 620, 2023
|
|
8Z2O
 
 | | Crystal structure of 5-N-alpha-glycinylthymidine (N-alpha-GlyT) FAD-dependent lyase gp47/NGTO from Pseudomonads phage PaMx11 | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, FLAVIN-ADENINE DINUCLEOTIDE, Flavin-dependent lyase, ... | | Authors: | Wen, Y, Guo, W.T, Wu, B.X. | | Deposit date: | 2024-04-13 | | Release date: | 2024-10-02 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural insights into the biosynthetic mechanism of N alpha-GlyT and 5-NmdU hypermodifications of DNA.
Nucleic Acids Res., 52, 2024
|
|
8Z2M
 
 | |
7WW3
 
 | |
8YHH
 
 | | The Crystal Structure of Mitotic Kinesin Eg5 from Biortus | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Wang, F, Cheng, W, Lv, Z, Qi, J, Wu, B. | | Deposit date: | 2024-02-28 | | Release date: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | The Crystal Structure of Mitotic Kinesin Eg5 from Biortus
To Be Published
|
|
7WW2
 
 | | Structure of an Isocytosine specific deaminase Vcz | | Descriptor: | 8-oxoguanine deaminase, ZINC ION | | Authors: | Li, X.J, Wu, B.X. | | Deposit date: | 2022-02-12 | | Release date: | 2023-02-22 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural characterization of an isocytosine-specific deaminase VCZ reveals its application potential in the anti-cancer therapy.
Iscience, 26, 2023
|
|
8Z79
 
 | |
