8T3Y
 
 | | Structure of Bre1-nucleosome complex - state1 | | Descriptor: | 601 DNA Strand 1, 601 DNA Strand 2, E3 ubiquitin-protein ligase BRE1, ... | | Authors: | Zhao, F, Hicks, C.W, Wolberger, C. | | Deposit date: | 2023-06-08 | | Release date: | 2023-10-18 | | Last modified: | 2023-11-22 | | Method: | ELECTRON MICROSCOPY (3.47 Å) | | Cite: | Mechanism of histone H2B monoubiquitination by Bre1.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8G6Q
 
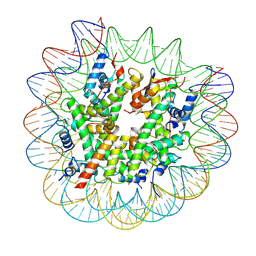 | | H2AK119ub-modified nucleosome ubiquitin position 1 | | Descriptor: | 601 DNA (147-MER), Histone H2A, Histone H2B 1.1, ... | | Authors: | Hicks, C.W, Wolberger, C, Keogh, M. | | Deposit date: | 2023-02-15 | | Release date: | 2024-02-21 | | Last modified: | 2024-09-25 | | Method: | ELECTRON MICROSCOPY (3.41 Å) | | Cite: | Ubiquitinated histone H2B as gatekeeper of the nucleosome acidic patch.
Nucleic Acids Res., 52, 2024
|
|
8G6S
 
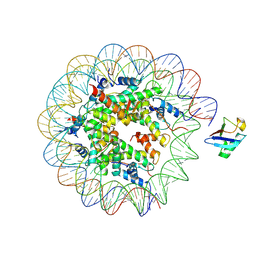 | | H2AK119ub-modified nucleosome ubiquitin position 2 | | Descriptor: | 601 DNA (147-MER), Histone H2A, Histone H2B 1.1, ... | | Authors: | Hicks, C.W, Wolberger, C, Keogh, M. | | Deposit date: | 2023-02-15 | | Release date: | 2024-02-21 | | Last modified: | 2024-09-25 | | Method: | ELECTRON MICROSCOPY (3.47 Å) | | Cite: | Ubiquitinated histone H2B as gatekeeper of the nucleosome acidic patch.
Nucleic Acids Res., 52, 2024
|
|
8G6G
 
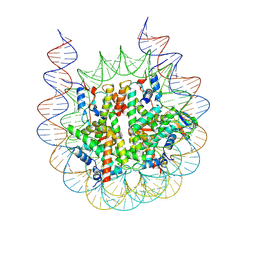 | | H2BK120ub+H3K79me2-modified nucleosome ubiquitin position 5 | | Descriptor: | 601 DNA (185-MER), Histone H2A, Histone H2B 1.1, ... | | Authors: | Hicks, C.W, Wolberger, C, Keogh, M. | | Deposit date: | 2023-02-15 | | Release date: | 2024-02-21 | | Last modified: | 2024-09-25 | | Method: | ELECTRON MICROSCOPY (2.93 Å) | | Cite: | Ubiquitinated histone H2B as gatekeeper of the nucleosome acidic patch.
Nucleic Acids Res., 52, 2024
|
|
4R7E
 
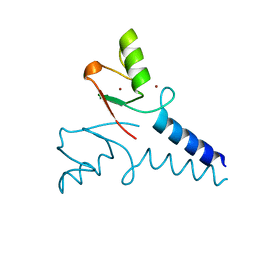 | | Structure of Bre1 RING domain | | Descriptor: | E3 ubiquitin-protein ligase BRE1, ZINC ION | | Authors: | Kumar, P, Wolberger, C. | | Deposit date: | 2014-08-27 | | Release date: | 2015-05-13 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.251 Å) | | Cite: | Structure of the yeast Bre1 RING domain.
Proteins, 83, 2015
|
|
2CRO
 
 | |
3MHH
 
 | | Structure of the SAGA Ubp8/Sgf11/Sus1/Sgf73 DUB module | | Descriptor: | Protein SUS1, SAGA-associated factor 11, SAGA-associated factor 73, ... | | Authors: | Samara, N.L, Datta, A.B, Berndsen, C.E, Zhang, X, Yao, T, Cohen, R.E, Wolberger, C. | | Deposit date: | 2010-04-08 | | Release date: | 2010-04-21 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structural insights into the assembly and function of the SAGA deubiquitinating module.
Science, 328, 2010
|
|
3JR3
 
 | | Sir2 bound to acetylated peptide | | Descriptor: | Acetylated Peptide, NAD-dependent deacetylase, ZINC ION | | Authors: | Hawse, W.F, Wolberger, C. | | Deposit date: | 2009-09-08 | | Release date: | 2009-09-29 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure-based mechanism of ADP-ribosylation by sirtuins.
J.Biol.Chem., 284, 2009
|
|
1AKH
 
 | | MAT A1/ALPHA2/DNA TERNARY COMPLEX | | Descriptor: | DNA (5'-D(*TP*AP*CP*AP*TP*GP*TP*AP*AP*AP*AP*AP*TP*TP*TP*AP*C P*AP*TP*CP*A)-3'), DNA (5'-D(*TP*AP*TP*GP*AP*TP*GP*TP*AP*AP*AP*TP*TP*TP*TP*TP*A P*CP*AP*TP*G)-3'), PROTEIN (MATING-TYPE PROTEIN A-1), ... | | Authors: | Li, T, Jin, Y, Vershon, A.K, Wolberger, C. | | Deposit date: | 1997-05-19 | | Release date: | 1998-05-20 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of the MATa1/MATalpha2 homeodomain heterodimer in complex with DNA containing an A-tract.
Nucleic Acids Res., 26, 1998
|
|
3MHS
 
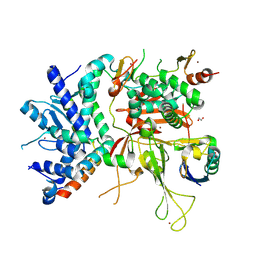 | | Structure of the SAGA Ubp8/Sgf11/Sus1/Sgf73 DUB module bound to ubiquitin aldehyde | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, Protein SUS1, ... | | Authors: | Samara, N.L, Datta, A.B, Berndsen, C.E, Zhang, X, Yao, T, Cohen, R.E, Wolberger, C. | | Deposit date: | 2010-04-08 | | Release date: | 2010-04-21 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Structural insights into the assembly and function of the SAGA deubiquitinating module.
Science, 328, 2010
|
|
3HM3
 
 | |
6NOG
 
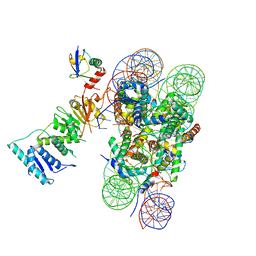 | | Poised-state Dot1L bound to the H2B-Ubiquitinated nucleosome | | Descriptor: | 601 DNA Strand 1, 601 DNA Strand 2, Histone H2A type 1, ... | | Authors: | Worden, E.J, Hoffmann, N.A, Wolberger, C. | | Deposit date: | 2019-01-16 | | Release date: | 2019-02-20 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Mechanism of Cross-talk between H2B Ubiquitination and H3 Methylation by Dot1L.
Cell, 176, 2019
|
|
6NJ9
 
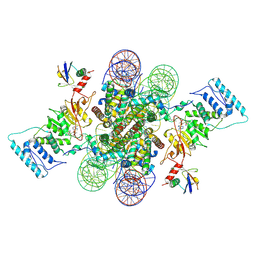 | | Active state Dot1L bound to the H2B-Ubiquitinated nucleosome, 2-to-1 complex | | Descriptor: | 601 DNA Strand 1, 601 DNA Strand 2, Histone H2A type 1, ... | | Authors: | Worden, E.J, Hoffmann, N.A, Wolberger, C. | | Deposit date: | 2019-01-02 | | Release date: | 2019-02-20 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.96 Å) | | Cite: | Mechanism of Cross-talk between H2B Ubiquitination and H3 Methylation by Dot1L.
Cell, 176, 2019
|
|
6NQA
 
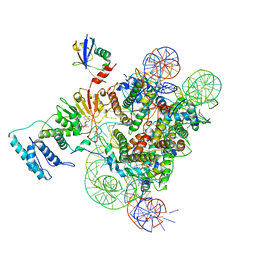 | | Active state Dot1L bound to the H2B-Ubiquitinated nucleosome, 1-to-1 complex | | Descriptor: | 601 DNA Strand 1, 601 DNA Strand 2, Histone H2A type 1, ... | | Authors: | Worden, E.J, Hoffmann, N.A, Wolberger, C. | | Deposit date: | 2019-01-19 | | Release date: | 2019-02-20 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.54 Å) | | Cite: | Mechanism of Cross-talk between H2B Ubiquitination and H3 Methylation by Dot1L.
Cell, 176, 2019
|
|
4DHZ
 
 | | The structure of h/ceOTUB1-ubiquitin aldehyde-UBC13~Ub | | Descriptor: | Ubiquitin, Ubiquitin aldehyde, Ubiquitin thioesterase otubain-like, ... | | Authors: | Wiener, R, Zhang, X, Wang, T, Wolberger, C. | | Deposit date: | 2012-01-30 | | Release date: | 2012-02-22 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (3.11 Å) | | Cite: | The mechanism of OTUB1-mediated inhibition of ubiquitination.
Nature, 483, 2012
|
|
8V25
 
 | |
8V28
 
 | |
8V26
 
 | |
8V27
 
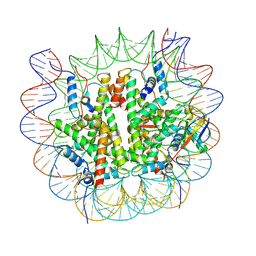 | |
1YC5
 
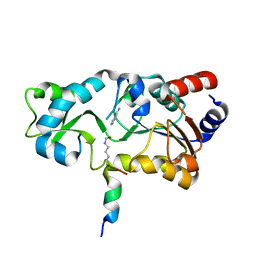 | | Sir2-p53 peptide-nicotinamide | | Descriptor: | Cellular tumor antigen p53 peptide, NAD-dependent deacetylase, NICOTINAMIDE, ... | | Authors: | Avalos, J.L, Bever, M.K, Wolberger, C. | | Deposit date: | 2004-12-21 | | Release date: | 2005-04-26 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Mechanism of sirtuin inhibition by nicotinamide: altering the NAD(+) cosubstrate specificity of a Sir2 enzyme.
Mol.Cell, 17, 2005
|
|
4DHI
 
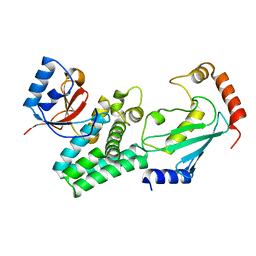 | | Structure of C. elegans OTUB1 bound to human UBC13 | | Descriptor: | Ubiquitin thioesterase otubain-like, Ubiquitin-conjugating enzyme E2 N | | Authors: | Wiener, R, Zhang, X, Wang, T, Wolberger, C. | | Deposit date: | 2012-01-27 | | Release date: | 2012-02-22 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The mechanism of OTUB1-mediated inhibition of ubiquitination.
Nature, 483, 2012
|
|
1YRN
 
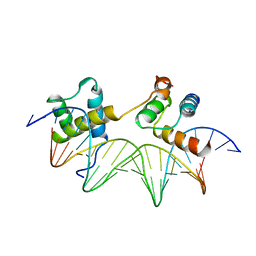 | | CRYSTAL STRUCTURE OF THE MATA1/MATALPHA2 HOMEODOMAIN HETERODIMER BOUND TO DNA | | Descriptor: | DNA (5'-D(*TP*AP*CP*AP*TP*GP*TP*AP*AP*TP*TP*TP*AP*TP*TP*AP*C P*AP*TP*CP*A)-3'), DNA (5'-D(*TP*AP*TP*GP*AP*TP*GP*TP*AP*AP*TP*AP*AP*AP*TP*TP*A P*CP*AP*TP*G)-3'), PROTEIN (MAT A1 HOMEODOMAIN), ... | | Authors: | Li, T, Stark, M.R, Johnson, A.D, Wolberger, C. | | Deposit date: | 1995-11-02 | | Release date: | 1996-01-29 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of the MATa1/MAT alpha 2 homeodomain heterodimer bound to DNA.
Science, 270, 1995
|
|
4DHJ
 
 | | The structure of a ceOTUB1 ubiquitin aldehyde UBC13~Ub complex | | Descriptor: | Ubiquitin, Ubiquitin aldehyde, Ubiquitin thioesterase otubain-like, ... | | Authors: | Wiener, R, Zhang, X, Wang, T, Wolberger, C. | | Deposit date: | 2012-01-27 | | Release date: | 2012-02-22 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | The mechanism of OTUB1-mediated inhibition of ubiquitination.
Nature, 483, 2012
|
|
3D4B
 
 | | Crystal structure of Sir2Tm in complex with Acetyl p53 peptide and DADMe-NAD+ | | Descriptor: | 5'-O-[(R)-{[(R)-{[(3R,4R)-1-(3-carbamoylbenzyl)-4-hydroxypyrrolidin-3-yl]methoxy}(hydroxy)phosphoryl]methyl}(hydroxy)phosphoryl]adenosine, Acetyl P53 peptide, NAD-dependent deacetylase, ... | | Authors: | Hawse, W.F, Hoff, K.G, Fatkins, D, Daines, A, Zubkova, O.V, Schramm, V.L, Zheng, W, Wolberger, C. | | Deposit date: | 2008-05-14 | | Release date: | 2008-09-30 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural insights into intermediate steps in the Sir2 deacetylation reaction.
Structure, 16, 2008
|
|
4FIP
 
 | | Structure of the SAGA Ubp8(S144N)/Sgf11(1-72, Delta-ZnF)/Sus1/Sgf73 DUB module | | Descriptor: | Protein SUS1, SAGA-associated factor 11, SAGA-associated factor 73, ... | | Authors: | Samara, N.L, Ringel, A.E, Wolberger, C. | | Deposit date: | 2012-06-10 | | Release date: | 2012-07-25 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.686 Å) | | Cite: | A Role for Intersubunit Interactions in Maintaining SAGA Deubiquitinating Module Structure and Activity.
Structure, 20, 2012
|
|
