4URV
 
 | | The crystal structure of H-Ras and SOS in complex with ligands | | Descriptor: | 4-(4-BROMOPHENYL)PIPERIDIN-4-OL, FORMIC ACID, GTPASE HRAS, ... | | Authors: | Winter, J.J.G, Anderson, M, Blades, K, Brassington, C, Breeze, A.L, Chresta, C, Embrey, K, Fairley, G, Faulder, P, Finlay, M.R.V, Kettle, J.G, Nowak, T, Overman, R, Patel, S.J, Perkins, P, Spadola, L, Tart, J, Tucker, J, Wrigley, G. | | Deposit date: | 2014-07-02 | | Release date: | 2015-03-04 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.58 Å) | | Cite: | Small Molecule Binding Sites on the Ras:SOS Complex Can be Exploited for Inhibition of Ras Activation.
J.Med.Chem., 58, 2015
|
|
4US1
 
 | | The crystal structure of H-Ras and SOS in complex with ligands | | Descriptor: | (3S)-3-[3-(aminomethyl)phenyl]-1-ethylpyrrolidine-2,5-dione, GTPASE HRAS, SON OF SEVENLESS HOMOLOG 1 | | Authors: | Winter, J.J.G, Anderson, M, Blades, K, Brassington, C, Breeze, A.L, Chresta, C, Embrey, K, Fairley, G, Faulder, P, Finlay, M.R.V, Kettle, J.G, Nowak, T, Overman, R, Patel, S.J, Perkins, P, Spadola, L, Tart, J, Tucker, J, Wrigley, G. | | Deposit date: | 2014-07-02 | | Release date: | 2015-03-04 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Small Molecule Binding Sites on the Ras:SOS Complex Can be Exploited for Inhibition of Ras Activation.
J.Med.Chem., 58, 2015
|
|
4URW
 
 | | The crystal structure of H-Ras and SOS in complex with ligands | | Descriptor: | 2-(2,6-DIMETHYLPHENYL)-4-(METHYLSULFANYL)-6-(PIPERAZIN-1-YL)-1,3,5-TRIAZINE, GTPASE HRAS, SON OF SEVENLESS HOMOLOG 1 | | Authors: | Winter, J.J.G, Anderson, M, Blades, K, Brassington, C, Breeze, A.L, Chresta, C, Embrey, K, Fairley, G, Faulder, P, Finlay, M.R.V, Kettle, J.G, Nowak, T, Overman, R, Patel, S.J, Perkins, P, Spadola, L, Tart, J, Tucker, J, Wrigley, G. | | Deposit date: | 2014-07-02 | | Release date: | 2015-03-04 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.76 Å) | | Cite: | Small Molecule Binding Sites on the Ras:SOS Complex Can be Exploited for Inhibition of Ras Activation.
J.Med.Chem., 58, 2015
|
|
4URY
 
 | | The crystal structure of H-Ras and SOS in complex with ligands | | Descriptor: | GTPASE HRAS, N-[(4-aminophenyl)sulfonyl]cyclopropanecarboxamide, SON OF SEVENLESS HOMOLOG 1 | | Authors: | Winter, J.J.G, Anderson, M, Blades, K, Brassington, C, Breeze, A.L, Chresta, C, Embrey, K, Fairley, G, Faulder, P, Finlay, M.R.V, Kettle, J.G, Nowak, T, Overman, R, Patel, S.J, Perkins, P, Spadola, L, Tart, J, Tucker, J, Wrigley, G. | | Deposit date: | 2014-07-02 | | Release date: | 2015-03-04 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.47 Å) | | Cite: | Small Molecule Binding Sites on the Ras:SOS Complex Can be Exploited for Inhibition of Ras Activation.
J.Med.Chem., 58, 2015
|
|
4US0
 
 | | The crystal structure of H-Ras and SOS in complex with ligands | | Descriptor: | 1-ETHYL-PYRROLIDINE-2,5-DIONE, GTPase HRas, Son of sevenless homolog 1 | | Authors: | Winter, J.J.G, Anderson, M, Blades, K, Brassington, C, Breeze, A.L, Chresta, C, Embrey, K, Fairley, G, Faulder, P, Finlay, M.R.V, Kettle, J.G, Nowak, T, Overman, R, Patel, S.J, Perkins, P, Spadola, L, Tart, J, Tucker, J, Wrigley, G. | | Deposit date: | 2014-07-02 | | Release date: | 2015-03-04 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Small Molecule Binding Sites on the Ras:SOS Complex Can be Exploited for Inhibition of Ras Activation.
J.Med.Chem., 58, 2015
|
|
4URZ
 
 | | The crystal structure of H-Ras and SOS in complex with ligands | | Descriptor: | 1-[(4-aminophenyl)sulfonyl]piperidin-2-one, GTPASE HRAS, SON OF SEVENLESS HOMOLOG 1 | | Authors: | Winter, J.J.G, Anderson, M, Blades, K, Brassington, C, Breeze, A.L, Chresta, C, Embrey, K, Fairley, G, Faulder, P, Finlay, M.R.V, Kettle, J.G, Nowak, T, Overman, R, Patel, S.J, Perkins, P, Spadola, L, Tart, J, Tucker, J, Wrigley, G. | | Deposit date: | 2014-07-02 | | Release date: | 2015-03-04 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Small Molecule Binding Sites on the Ras:SOS Complex Can be Exploited for Inhibition of Ras Activation.
J.Med.Chem., 58, 2015
|
|
4URX
 
 | | The crystal structure of H-Ras and SOS in complex with ligands | | Descriptor: | 1-(4-bromobenzyl)pyrrolidine, 6-bromo-1H-indole, FORMIC ACID, ... | | Authors: | Winter, J.J.G, Anderson, M, Blades, K, Brassington, C, Breeze, A.L, Chresta, C, Embrey, K, Fairley, G, Faulder, P, Finlay, M.R.V, Kettle, J.G, Nowak, T, Overman, R, Patel, S.J, Perkins, P, Spadola, L, Tart, J, Tucker, J, Wrigley, G. | | Deposit date: | 2014-07-02 | | Release date: | 2015-03-04 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Small Molecule Binding Sites on the Ras:SOS Complex Can be Exploited for Inhibition of Ras Activation.
J.Med.Chem., 58, 2015
|
|
4URU
 
 | | The crystal structure of H-Ras and SOS in complex with ligands | | Descriptor: | 4-METHOXY-N-(1,3-THIAZOL-2-YL)BENZENESULFONAMIDE, GTPASE HRAS, SON OF SEVENLESS HOMOLOG 1 | | Authors: | Winter, J.J.G, Anderson, M, Blades, K, Brassington, C, Breeze, A.L, Chresta, C, Embrey, K, Fairley, G, Faulder, P, Finlay, M.R.V, Kettle, J.G, Nowak, T, Overman, R, Patel, S.J, Perkins, P, Spadola, L, Tart, J, Tucker, J, Wrigley, G. | | Deposit date: | 2014-07-02 | | Release date: | 2015-03-04 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.83 Å) | | Cite: | Small Molecule Binding Sites on the Ras:SOS Complex Can be Exploited for Inhibition of Ras Activation.
J.Med.Chem., 58, 2015
|
|
4RO5
 
 | | Crystal structure of the SAT domain from the non-reducing fungal polyketide synthase CazM | | Descriptor: | GLYCEROL, SAT domain from CazM | | Authors: | Winter, J.M, Cascio, D, Sawaya, M.R, Tang, Y. | | Deposit date: | 2014-10-27 | | Release date: | 2015-09-09 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Biochemical and Structural Basis for Controlling Chemical Modularity in Fungal Polyketide Biosynthesis.
J.Am.Chem.Soc., 137, 2015
|
|
4RPM
 
 | | Crystal structure of the SAT domain from the non-reducing fungal polyketide synthase CazM with bound hexanoyl | | Descriptor: | HEXANOIC ACID, HEXANOYL-COENZYME A, SAT domain from CazM | | Authors: | Winter, J.M, Cascio, D, Sawaya, M.R, Tang, Y. | | Deposit date: | 2014-10-30 | | Release date: | 2015-09-09 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Biochemical and Structural Basis for Controlling Chemical Modularity in Fungal Polyketide Biosynthesis.
J.Am.Chem.Soc., 137, 2015
|
|
4US2
 
 | | The crystal structure of H-Ras and SOS in complex with ligands | | Descriptor: | 3-[(3R)-1-ethyl-2,5-dioxopyrrolidin-3-yl]benzamide, GTPASE HRAS, SON OF SEVENLESS HOMOLOG 1 | | Authors: | Winter, J.J.G, Anderson, M, Blades, K, Brassington, C, Breeze, A.L, Chresta, C, Embrey, K, Fairley, G, Faulder, P, Finlay, M.R.V, Kettle, J.G, Nowak, T, Overman, R, Patel, S.J, Perkins, P, Spadola, L, Tart, J, Tucker, J, Wrigley, G. | | Deposit date: | 2014-07-02 | | Release date: | 2015-03-04 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Small Molecule Binding Sites on the Ras:SOS Complex Can be Exploited for Inhibition of Ras Activation.
J.Med.Chem., 58, 2015
|
|
1HCO
 
 | |
3IFV
 
 | | Crystal structure of the Haloferax volcanii proliferating cell nuclear antigen | | Descriptor: | PCNA, SODIUM ION | | Authors: | Winter, J.A, Christofi, P, Morroll, S, Bunting, K.A. | | Deposit date: | 2009-07-26 | | Release date: | 2009-09-01 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The crystal structure of Haloferax volcanii proliferating cell nuclear antigen reveals unique surface charge characteristics due to halophilic adaptation
Bmc Struct.Biol., 9, 2009
|
|
3ET6
 
 | | The crystal structure of the catalytic domain of a eukaryotic guanylate cyclase | | Descriptor: | PHOSPHATE ION, Soluble guanylyl cyclase beta | | Authors: | Winger, J.A, Derbyshire, E.R, Lamers, M.H, Marletta, M.A, Kuriyan, J. | | Deposit date: | 2008-10-07 | | Release date: | 2008-10-14 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | The crystal structure of the catalytic domain of a eukaryotic guanylate cyclase.
Bmc Struct.Biol., 8, 2008
|
|
3FW1
 
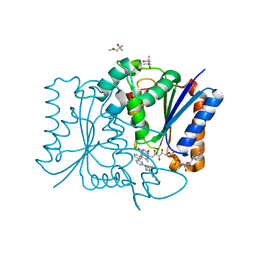 | | Quinone Reductase 2 | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, 4-(4-METHYL-PIPERAZIN-1-YLMETHYL)-N-[4-METHYL-3-(4-PYRIDIN-3-YL-PYRIMIDIN-2-YLAMINO)-PHENYL]-BENZAMIDE, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Winger, J.A, Hantschel, O, Superti-Furga, G, Kuriyan, J. | | Deposit date: | 2009-01-16 | | Release date: | 2009-03-10 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | The structure of the leukemia drug imatinib bound to human quinone reductase 2 (NQO2).
Bmc Struct.Biol., 9, 2009
|
|
4P3F
 
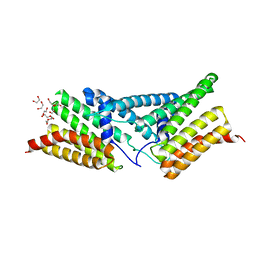 | | Structure of the human SRP68-RBD | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, Signal recognition particle subunit SRP68 | | Authors: | Grotwinkel, J.T, Wild, K, Sinning, I. | | Deposit date: | 2014-03-07 | | Release date: | 2014-04-16 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.699 Å) | | Cite: | SRP RNA remodeling by SRP68 explains its role in protein translocation.
Science, 344, 2014
|
|
4P3E
 
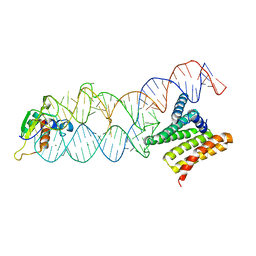 | | Structure of the human SRP S domain | | Descriptor: | MAGNESIUM ION, SRP RNA (124-mer), Signal recognition particle 19 kDa protein, ... | | Authors: | Grotwinkel, J.T, Wild, K, Sinning, I. | | Deposit date: | 2014-03-07 | | Release date: | 2014-04-16 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | SRP RNA remodeling by SRP68 explains its role in protein translocation.
Science, 344, 2014
|
|
4P3G
 
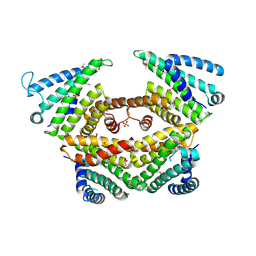 | |
2HCO
 
 | |
2H15
 
 | | Carbonic anhydrase inhibitors: Clashing with Ala65 as a means of designing isozyme-selective inhibitors that show low affinity for the ubiquitous isozyme II | | Descriptor: | Carbonic anhydrase 2, MERCURY (II) ION, N-{[(3AS,5AR,8AR,8BS)-2,2,7,7-TETRAMETHYLTETRAHYDRO-3AH-BIS[1,3]DIOXOLO[4,5-B:4',5'-D]PYRAN-3A-YL]METHYL}SULFAMIDE, ... | | Authors: | Winum, J.Y, Temperini, C, Ciattini, S, Scozzafava, A, Supuran, C.T. | | Deposit date: | 2006-05-16 | | Release date: | 2007-03-27 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Carbonic anhydrase inhibitors: clash with Ala65 as a means for designing inhibitors with low affinity for the ubiquitous isozyme II, exemplified by the crystal structure of the topiramate sulfamide analogue.
J.Med.Chem., 49, 2006
|
|
2LQT
 
 | |
3ZR8
 
 | | Crystal structure of RxLR effector Avr3a11 from Phytophthora capsici | | Descriptor: | AVR3A11, CHLORIDE ION, TRIETHYLENE GLYCOL | | Authors: | Boutemy, L.S, King, S.R.F, Win, J, Hughes, R.K, Clarke, T.A, Blumenschein, T.M.A, Kamoun, S, Banfield, M.J. | | Deposit date: | 2011-06-15 | | Release date: | 2011-08-03 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (0.9 Å) | | Cite: | Structures of Phytophthora Rxlr Effector Proteins: A Conserved But Adaptable Fold Underpins Functional Diversity.
J.Biol.Chem., 286, 2011
|
|
3ZRG
 
 | | Crystal structure of RxLR effector PexRD2 from Phytophthora infestans | | Descriptor: | BROMIDE ION, PEXRD2 FAMILY SECRETED RXLR EFFECTOR PEPTIDE, PUTATIVE | | Authors: | King, S.R.F, Boutemy, L.S, Win, J, Hughes, R.K, Clarke, T.A, Blumenschein, T.M.A, Kamoun, S, Banfield, M.J. | | Deposit date: | 2011-06-16 | | Release date: | 2011-08-03 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structures of Phytophthora Rxlr Effector Proteins: A Conserved But Adaptable Fold Underpins Functional Diversity.
J.Biol.Chem., 286, 2011
|
|
1W3B
 
 | | The superhelical TPR domain of O-linked GlcNAc transferase reveals structural similarities to importin alpha. | | Descriptor: | CALCIUM ION, UDP-N-ACETYLGLUCOSAMINE--PEPTIDE N-ACETYLGLUCOSAMINYLTRANSFERASE 110 | | Authors: | Jinek, M, Rehwinkel, J, Lazarus, B.D, Izaurralde, E, Hanover, J.A, Conti, E. | | Deposit date: | 2004-07-14 | | Release date: | 2004-09-09 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | The Superhelical Tpr-Repeat Domain of O-Linked Glcnac Transferase Exhibits Structural Similarities to Importin Alpha
Nat.Struct.Mol.Biol., 11, 2004
|
|
3K96
 
 | | 2.1 Angstrom resolution crystal structure of glycerol-3-phosphate dehydrogenase (gpsA) from Coxiella burnetii | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, BETA-MERCAPTOETHANOL, Glycerol-3-phosphate dehydrogenase [NAD(P)+] | | Authors: | Minasov, G, Halavaty, A, Shuvalova, L, Dubrovska, I, Winsor, J, Peterson, S.N, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2009-10-15 | | Release date: | 2009-10-27 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | 2.1 Angstrom Resolution Crystal Structure of Glycerol-3-phosphate Dehydrogenase (gpsA) from Coxiella burnetii.
TO BE PUBLISHED
|
|
