2WF7
 
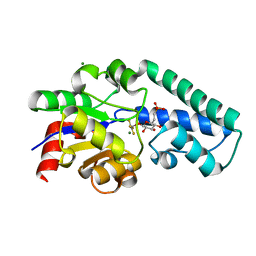 | | Structure of Beta-Phosphoglucomutase inhibited with Glucose-6- phosphonate and Aluminium tetrafluoride | | Descriptor: | 6,7-dideoxy-7-phosphono-beta-D-gluco-heptopyranose, BETA-PHOSPHOGLUCOMUTASE, MAGNESIUM ION, ... | | Authors: | Bowler, M.W, Baxter, N.J, Webster, C.E, Pollard, S, Alizadeh, T, Hounslow, A.M, Cliff, M.J, Bermel, W, Williams, N.H, Hollfelder, F, Blackburn, G.M, Waltho, J.P. | | Deposit date: | 2009-04-03 | | Release date: | 2010-05-19 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Alpha-Fluorophosphonates Reveal How a Phosphomutase Conserves Transition State Conformation Over Hexose Recognition in its Two-Step Reaction.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
2WFA
 
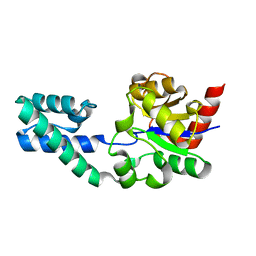 | | Structure of Beta-Phosphoglucomutase inhibited with Beryllium trifluoride, in an open conformation. | | Descriptor: | BERYLLIUM TRIFLUORIDE ION, BETA-PHOSPHOGLUCOMUTASE, MAGNESIUM ION | | Authors: | Bowler, M.W, Baxter, N.J, Webster, C.E, Pollard, S, Alizadeh, T, Hounslow, A.M, Cliff, M.J, Bermel, W, Williams, N.H, Hollfelder, F, Blackburn, G.M, Waltho, J.P. | | Deposit date: | 2009-04-03 | | Release date: | 2010-05-26 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Near Attack Conformers Dominate Beta-Phosphoglucomutase Complexes Where Geometry and Charge Distribution Reflect Those of Substrate.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
2WF8
 
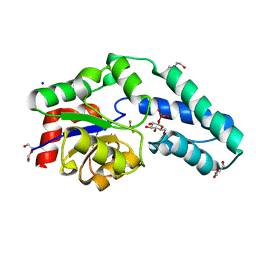 | | Structure of Beta-Phosphoglucomutase inhibited with Glucose-6- phosphate, Glucose-1-phosphate and Beryllium trifluoride | | Descriptor: | 1-O-phosphono-alpha-D-glucopyranose, 6-O-phosphono-beta-D-glucopyranose, BERYLLIUM TRIFLUORIDE ION, ... | | Authors: | Bowler, M.W, Baxter, N.J, Webster, C.E, Pollard, S, Alizadeh, T, Hounslow, A.M, Cliff, M.J, Bermel, W, Williams, N.H, Hollfelder, F, Blackburn, G.M, Waltho, J.P. | | Deposit date: | 2009-04-03 | | Release date: | 2010-05-26 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Near attack conformers dominate beta-phosphoglucomutase complexes where geometry and charge distribution reflect those of substrate.
Proc. Natl. Acad. Sci. U.S.A., 109, 2012
|
|
2WF9
 
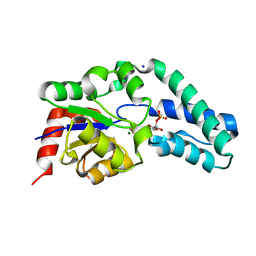 | | Structure of Beta-Phosphoglucomutase inhibited with Glucose-6- phosphate, and Beryllium trifluoride, crystal form 2 | | Descriptor: | 6-O-phosphono-alpha-D-glucopyranose, 6-O-phosphono-beta-D-glucopyranose, BERYLLIUM TRIFLUORIDE ION, ... | | Authors: | Bowler, M.W, Baxter, N.J, Webster, C.E, Pollard, S, Alizadeh, T, Hounslow, A.M, Cliff, M.J, Bermel, W, Williams, N.H, Hollfelder, F, Blackburn, G.M, Waltho, J.P. | | Deposit date: | 2009-04-03 | | Release date: | 2010-05-26 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Near Attack Conformers Dominate Beta-Phosphoglucomutase Complexes Where Geometry and Charge Distribution Reflect Those of Substrate.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3QPL
 
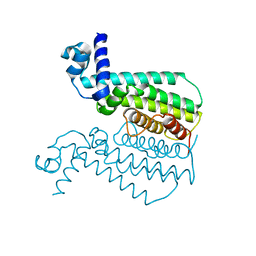 | | G106W mutant of EthR from Mycobacterium tuberculosis | | Descriptor: | HTH-type transcriptional regulator EthR | | Authors: | Carette, X, Willery, E, Hoos, S, Lecat-Guillet, N, Frenois, F, Dirie, B, Villeret, V, England, P, Deprez, B, Locht, C, Willand, N, Baulard, A. | | Deposit date: | 2011-02-14 | | Release date: | 2011-12-28 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural activation of the transcriptional repressor EthR from Mycobacterium tuberculosis by single amino acid change mimicking natural and synthetic ligands.
Nucleic Acids Res., 40, 2012
|
|
4V2I
 
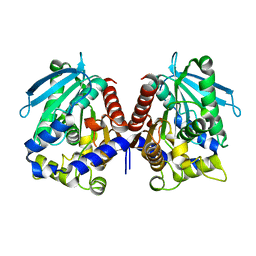 | | Biochemical characterization and structural analysis of a new cold- active and salt tolerant esterase from the marine bacterium Thalassospira sp | | Descriptor: | ESTERASE/LIPASE, MAGNESIUM ION | | Authors: | Santi, C.D, Leiros, H.-K.S, Scala, A.D, Pascale, D.D, Altermark, B, Willassen, N.-P. | | Deposit date: | 2014-10-10 | | Release date: | 2016-01-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.686 Å) | | Cite: | Biochemical Characterization and Structural Analysis of a New Cold-Active and Salt-Tolerant Esterase from the Marine Bacterium Thalassospira Sp.
Extremophiles, 20, 2016
|
|
2STB
 
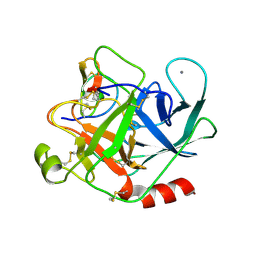 | | ANIONIC SALMON TRYPSIN IN COMPLEX WITH SQUASH SEED INHIBITOR (CUCURBITA PEPO TRYPSIN INHIBITOR II) | | Descriptor: | CALCIUM ION, PROTEIN (TRYPSIN INHIBITOR), PROTEIN (TRYPSIN) | | Authors: | Helland, R, Berglund, G.I, Otlewski, J, Apostoluk, W, Andersen, O.A, Willassen, N.P, Smalas, A.O. | | Deposit date: | 1998-12-11 | | Release date: | 2000-01-19 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | High-resolution structures of three new trypsin-squash-inhibitor complexes: a detailed comparison with other trypsins and their complexes.
Acta Crystallogr.,Sect.D, 55, 1999
|
|
2STA
 
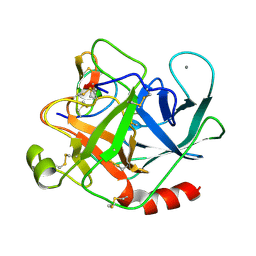 | | ANIONIC SALMON TRYPSIN IN COMPLEX WITH SQUASH SEED INHIBITOR (CUCURBITA MAXIMA TRYPSIN INHIBITOR I) | | Descriptor: | CALCIUM ION, PROTEIN (TRYPSIN INHIBITOR), PROTEIN (TRYPSIN) | | Authors: | Helland, R, Berglund, G.I, Otlewski, J, Apostoluk, W, Andersen, O.A, Willassen, N.P, Smalas, A.O. | | Deposit date: | 1998-12-10 | | Release date: | 2000-01-19 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | High-resolution structures of three new trypsin-squash-inhibitor complexes: a detailed comparison with other trypsins and their complexes.
Acta Crystallogr.,Sect.D, 55, 1999
|
|
2VND
 
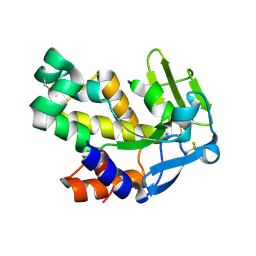 | | The N69Q mutant of Vibrio cholerae endonuclease I | | Descriptor: | CHLORIDE ION, ENDONUCLEASE I, MAGNESIUM ION | | Authors: | Niiranen, L, Altermark, B, Brandsdal, B.O, Leiros, H.-K.S, Helland, R, Smalas, A.O, Willassen, N.P. | | Deposit date: | 2008-02-04 | | Release date: | 2008-02-12 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Effects of Salt on the Kinetics and Thermodynamic Stability of Endonuclease I from Vibrio Salmonicida and Vibrio Cholerae.
FEBS J., 275, 2008
|
|
2W7W
 
 | |
8RCW
 
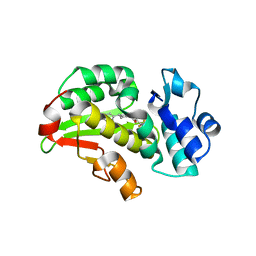 | | Crystal structure of the Mycobacterium tuberculosis regulator VirS (N-terminal fragment 4-208) in complex with the lead compound SMARt751 | | Descriptor: | 4,4,4-tris(fluoranyl)-1-[4-(4-fluorophenyl)piperidin-1-yl]butan-1-one, HTH-type transcriptional regulator VirS | | Authors: | Grosse, C, Sigoillot, M, Megalizzi, V, Tanina, A, Willand, N, Baulard, A.R, Wintjens, R. | | Deposit date: | 2023-12-07 | | Release date: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (1.692 Å) | | Cite: | Crystal structure of the Mycobacterium tuberculosis VirS regulator reveals its interaction with the lead compound SMARt751.
J.Struct.Biol., 216, 2024
|
|
7QVZ
 
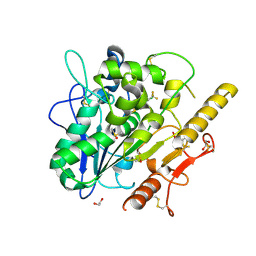 | | ARUK3001043_Notum | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, DIMETHYL SULFOXIDE, ... | | Authors: | Zhao, Y, Hillier, J, Willis, N.J, Mahy, W, Fish, P, Jones, E.Y. | | Deposit date: | 2022-01-24 | | Release date: | 2022-05-04 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Structure Activity Analysis of Notum Fragment Screen Hits with Development
To Be Published
|
|
3TKB
 
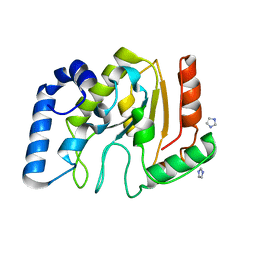 | | crystal structure of human uracil-DNA glycosylase D183G/K302R mutant | | Descriptor: | IMIDAZOLE, Uracil-DNA glycosylase | | Authors: | Assefa, N.G, Niiranen, L, Willassen, N.P, Smalas, A.O, Moe, E. | | Deposit date: | 2011-08-26 | | Release date: | 2011-10-12 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Thermal unfolding studies of cold adapted uracil-DNA N-glycosylase (UNG) from Atlantic cod (Gadus morhua). A comparative study with human UNG.
Comp.Biochem.Physiol. B: Biochem.Mol.Biol., 161, 2012
|
|
1BZX
 
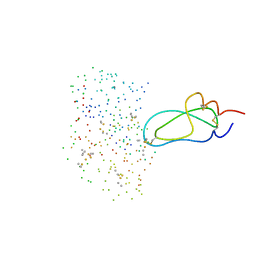 | | THE CRYSTAL STRUCTURE OF ANIONIC SALMON TRYPSIN IN COMPLEX WITH BOVINE PANCREATIC TRYPSIN INHIBITOR | | Descriptor: | CALCIUM ION, PROTEIN (BOVINE PANCREATIC TRYPSIN INHIBITOR), PROTEIN (TRYPSIN) | | Authors: | Helland, R, Leiros, I, Berglund, G.I, Willassen, N.P, Smalas, A.O. | | Deposit date: | 1998-11-05 | | Release date: | 1998-11-11 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The crystal structure of anionic salmon trypsin in complex with bovine pancreatic trypsin inhibitor.
Eur.J.Biochem., 256, 1998
|
|
1A0J
 
 | | CRYSTAL STRUCTURE OF A NON-PSYCHROPHILIC TRYPSIN FROM A COLD-ADAPTED FISH SPECIES. | | Descriptor: | BENZAMIDINE, CALCIUM ION, SULFATE ION, ... | | Authors: | Schroeder, H.-K, Willassen, N.P, Smalaas, A.O. | | Deposit date: | 1997-12-01 | | Release date: | 1999-01-13 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure of a non-psychrophilic trypsin from a cold-adapted fish species.
Acta Crystallogr.,Sect.D, 54, 1998
|
|
5ICJ
 
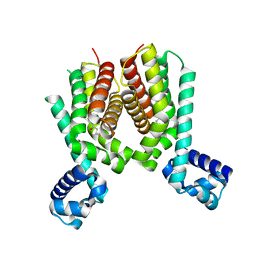 | | Crystal structure of the Mycobacterium tuberculosis transcriptional repressor EthR2 in complex with BDM41420 | | Descriptor: | 4,4,4-trifluoro-1-(3-phenyl-1-oxa-2,8-diazaspiro[4.5]dec-2-en-8-yl)butan-1-one, Probable transcriptional regulatory protein | | Authors: | Wohlkonig, A, Remaut, H, Tanina, A, Meyer, F, Willand, N, Baulard, A.R, Wintjens, R. | | Deposit date: | 2016-02-23 | | Release date: | 2017-04-26 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Reversion of antibiotic resistance in Mycobacterium tuberculosis by spiroisoxazoline SMARt-420.
Science, 355, 2017
|
|
3Q0V
 
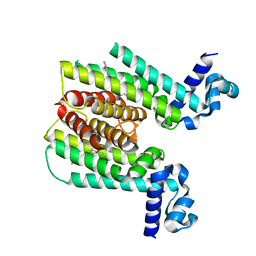 | | ETHR From mycobacterium tuberculosis in complex with compound bdm31369 | | Descriptor: | 4-methyl-1-{4-[3-(thiophen-2-yl)-1,2,4-oxadiazol-5-yl]piperidin-1-yl}pentan-1-one, HTH-type transcriptional regulator EthR | | Authors: | Flipo, M, Desroses, M, Dirie, B, Carette, X, Leroux, F, Lens, Z, Rucktooa, P, Leroux, F, Piveteau, C, Demirkaya, F, Locht, C, Villeret, V, Christophe, T, Jeon, H.K, Brodin, P, Deprez, B, Baulard, A, Willand, N. | | Deposit date: | 2010-12-16 | | Release date: | 2012-01-11 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Design, synthesis and optimization of new EthR inhibitors. A new alternative approach to fight tuberculosis by boosting ethionamide
To be Published
|
|
3Q0U
 
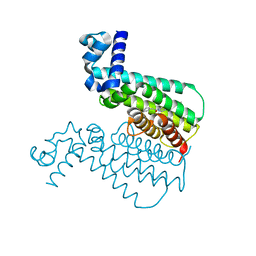 | | EthR from Mycobacterium tuberculosis in complex with compound BDM31379 | | Descriptor: | 2-phenyl-1-{4-[3-(thiophen-2-yl)-1,2,4-oxadiazol-5-yl]piperidin-1-yl}ethanone, HTH-type transcriptional regulator EthR | | Authors: | Flipo, M, Desroses, M, Dirie, B, Carette, X, Lens, Z, Rucktooa, P, Leroux, F, Piveteau, C, Demirkaya, F, Locht, C, Villeret, V, Christophe, T, Jeon, H.K, Brodin, P, Deprez, B, Baulard, A, Willand, N. | | Deposit date: | 2010-12-16 | | Release date: | 2012-01-11 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Design, synthesis and optimization of new EthR inhibitors. A new alternative approach to fight tuberculosis by boosting ethionamide
To be Published
|
|
3Q3S
 
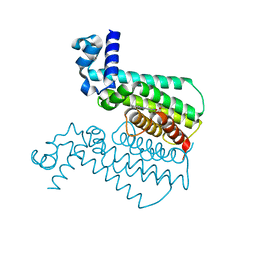 | |
3Q0W
 
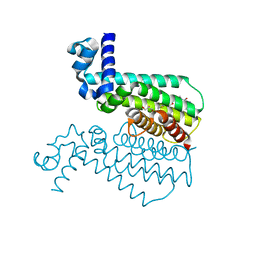 | | ETHR From mycobacterium tuberculosis in complex with compound BDM33066 | | Descriptor: | (2S)-2-amino-3-methyl-1-{4-[3-(thiophen-2-yl)-1,2,4-oxadiazol-5-yl]piperidin-1-yl}butan-1-one, GLYCEROL, HTH-type transcriptional regulator EthR | | Authors: | Flipo, M, Desrose, M, Dirie, B, Carette, X, Leroux, F, Lens, Z, Rucktooa, P, Piveteau, C, Demirkaya, F, Locht, C, Villeret, V, Christophe, T, Jeon, H.K, Brodin, P, Deprez, B, Baulard, A, Willand, N. | | Deposit date: | 2010-12-16 | | Release date: | 2011-12-21 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural activation of the transcriptional repressor EthR from Mycobacterium tuberculosis by single amino acid change mimicking natural and synthetic ligands.
Nucleic Acids Res., 40, 2012
|
|
4M3F
 
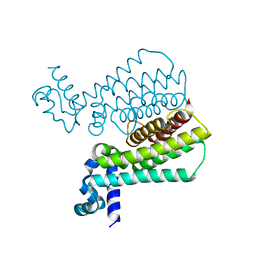 | | Rapid and efficient design of new inhibitors of Mycobacterium tuberculosis transcriptional repressor EthR using fragment growing, merging and linking approaches | | Descriptor: | HTH-type transcriptional regulator EthR, N-(3-methylbutyl)-4-(2-methyl-1,3-thiazol-4-yl)benzenesulfonamide | | Authors: | Villemagne, B, Flipo, M, Blondiaux, N, Crauste, C, Malaquin, S, Leroux, F, Piveteau, C, Villeret, V, Brodin, P, Villoutreix, B, Sperandio, O, Wohlkonig, A, Wintjens, R, Deprez, B, Baulard, A, Willand, N. | | Deposit date: | 2013-08-06 | | Release date: | 2014-06-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Ligand Efficiency Driven Design of New Inhibitors of Mycobacterium tuberculosis Transcriptional Repressor EthR Using Fragment Growing, Merging, and Linking Approaches.
J.Med.Chem., 57, 2014
|
|
4M3D
 
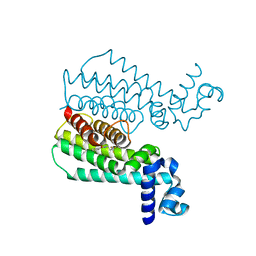 | | Rapid and efficient design of new inhibitors of Mycobacterium tuberculosis transcriptional repressor EthR using fragment growing, merging and linking approaches | | Descriptor: | 4-{3-[(phenylsulfonyl)amino]prop-1-yn-1-yl}-N-(3,3,3-trifluoropropyl)benzamide, HTH-type transcriptional regulator EthR | | Authors: | Villemagne, B, Flipo, M, Blondiaux, N, Crauste, C, Malaquin, S, Leroux, F, Piveteau, C, Villeret, V, Brodin, P, Villoutreix, B, Sperandio, O, Wohlkonig, A, Wintjens, R, Deprez, B, Baulard, A, Willand, N. | | Deposit date: | 2013-08-06 | | Release date: | 2014-06-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Ligand Efficiency Driven Design of New Inhibitors of Mycobacterium tuberculosis Transcriptional Repressor EthR Using Fragment Growing, Merging, and Linking Approaches.
J.Med.Chem., 57, 2014
|
|
4M3E
 
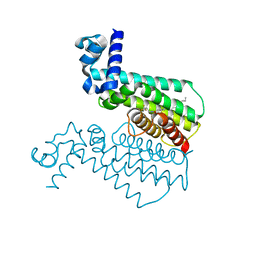 | | Rapid and efficient design of new inhibitors of Mycobacterium tuberculosis transcriptional repressor EthR using fragment growing, merging and linking approaches | | Descriptor: | 4-(2-{[(propylsulfonyl)amino]methyl}-1,3-thiazol-4-yl)-N-(3,3,3-trifluoropropyl)benzamide, HTH-type transcriptional regulator EthR | | Authors: | Villemagne, B, Flipo, M, Blondiaux, N, Crauste, C, Malaquin, S, Leroux, F, Piveteau, C, Villeret, V, Brodin, P, Villoutreix, B, Sperandio, O, Wohlkonig, A, Wintjens, R, Deprez, B, Baulard, A, Willand, N. | | Deposit date: | 2013-08-06 | | Release date: | 2014-06-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.109 Å) | | Cite: | Ligand Efficiency Driven Design of New Inhibitors of Mycobacterium tuberculosis Transcriptional Repressor EthR Using Fragment Growing, Merging, and Linking Approaches.
J.Med.Chem., 57, 2014
|
|
3SDG
 
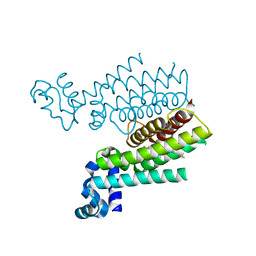 | | Ethionamide Boosters Part 2: Combining Bioisosteric Replacement and Structure-Based Drug Design to Solve Pharmacokinetic Issues in a Series of Potent 1,2,4-Oxadiazole EthR Inhibitors. | | Descriptor: | 4,4,4-trifluoro-1-{4-[3-(1,3-thiazol-2-yl)-1,2,4-oxadiazol-5-yl]piperidin-1-yl}butan-1-one, HTH-type transcriptional regulator EthR | | Authors: | Flipo, M, Desroses, M, Lecat-Guillet, N, Villemagne, B, Blondiaux, N, Leroux, F, Piveteau, C, Mathys, V, Flament, M.P, Siepmann, J, Villeret, V, Wohlkonig, A, Wintjens, R, Soror, S.H, Christophe, T, Jeon, H.K, Locht, C, Brodin, P, D prez, B, Baulard, A.R, Willand, N. | | Deposit date: | 2011-06-09 | | Release date: | 2011-12-07 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Ethionamide Boosters. 2. Combining Bioisosteric Replacement and Structure-Based Drug Design To Solve Pharmacokinetic Issues in a Series of Potent 1,2,4-Oxadiazole EthR Inhibitors.
J.Med.Chem., 55, 2012
|
|
4M3B
 
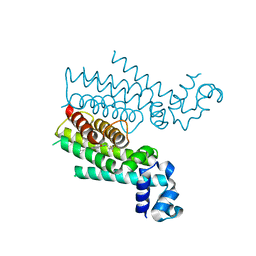 | | Rapid and efficient design of new inhibitors of Mycobacterium tuberculosis transcriptional repressor EthR using fragment growing, merging and linking approaches | | Descriptor: | 4-(2-methyl-1,3-thiazol-4-yl)-N-(3,3,3-trifluoropropyl)benzamide, HTH-type transcriptional regulator EthR | | Authors: | Villemagne, B, Flipo, M, Blondiaux, N, Crauste, C, Malaquin, S, Leroux, F, Piveteau, C, Villeret, V, Brodin, P, Villoutreix, B, Sperandio, O, Wohlkonig, A, Wintjens, R, Deprez, B, Baulard, A, Willand, N. | | Deposit date: | 2013-08-06 | | Release date: | 2014-06-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.001 Å) | | Cite: | Ligand Efficiency Driven Design of New Inhibitors of Mycobacterium tuberculosis Transcriptional Repressor EthR Using Fragment Growing, Merging, and Linking Approaches.
J.Med.Chem., 57, 2014
|
|
