1B05
 
 | |
1B3G
 
 | |
1B40
 
 | |
1B4Z
 
 | | OLIGO-PEPTIDE BINDING PROTEIN (OPPA) COMPLEXED WITH KDK | | Descriptor: | ACETATE ION, PROTEIN (OLIGO-PEPTIDE BINDING PROTEIN), PROTEIN (PEPTIDE LYS-ASP-LYS), ... | | Authors: | Tame, J.R.H, Sleigh, S.H, Wilkinson, A.J. | | Deposit date: | 1999-01-04 | | Release date: | 1999-01-13 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystallographic and calorimetric analysis of peptide binding to OppA protein.
J.Mol.Biol., 291, 1999
|
|
1OLC
 
 | |
4A95
 
 | | Plasmodium vivax N-myristoyltransferase with quinoline inhibitor | | Descriptor: | 2-oxopentadecyl-CoA, 3-(3-BUTYL-6-METHOXY-2-METHYL-QUINOLIN-4-YL)SULFANYLPROPANENITRILE, CHLORIDE ION, ... | | Authors: | Goncalves, V, Brannigan, J.A, Whalley, D, Ansell, K.H, Saxty, B, Holder, A.A, Wilkinson, A.J, Tate, E.W, Leatherbarrow, R.J. | | Deposit date: | 2011-11-24 | | Release date: | 2012-06-27 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Discovery of Plasmodium Vivax N-Myristoyltransferase Inhibitors: Screening, Synthesis, and Structural Characterization of Their Binding Mode.
J.Med.Chem., 55, 2012
|
|
4B14
 
 | | Plasmodium vivax N-myristoyltransferase with a bound benzofuran inhibitor (compound 26) | | Descriptor: | 2-oxopentadecyl-CoA, 3-methoxybenzyl 3-methyl-4-(piperidin-4-yloxy)-1-benzofuran-2-carboxylate, CHLORIDE ION, ... | | Authors: | Yu, Z, Brannigan, J.A, Moss, D.K, Brzozowski, A.M, Wilkinson, A.J, Holder, A.A, Tate, E.W, Leatherbarrow, R.J. | | Deposit date: | 2012-07-06 | | Release date: | 2012-10-17 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Design and Synthesis of Inhibitors of Plasmodium Falciparum N-Myristoyltransferase, a Promising Target for Anti-Malarial Drug Discovery.
J.Med.Chem., 55, 2012
|
|
4AXV
 
 | | Biochemical and structural characterization of the MpaA amidase as part of a conserved scavenging pathway for peptidoglycan derived peptides in gamma-proteobacteria | | Descriptor: | 1,2-ETHANEDIOL, MPAA, ZINC ION | | Authors: | Maqbool, A, Herve, M, Mengin-Lecreulx, D, Dodson, E, Wilkinson, A.J, Thomas, G.H. | | Deposit date: | 2012-06-14 | | Release date: | 2012-09-26 | | Last modified: | 2020-03-04 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Mpaa is a Murein-Tripeptide-Specific Zinc Carboxypeptidase that Functions as Part of a Catabolic Pathway for Peptidoglycan Derived Peptides in Gamma-Proteobacteria.
Biochem.J., 448, 2012
|
|
2YND
 
 | | Plasmodium vivax N-myristoyltransferase in complex with a pyrazole sulphonamide inhibitor. | | Descriptor: | 2,6-dichloro-4-(2-piperazin-1-ylpyridin-4-yl)-N-(1,3,5-trimethyl-1H-pyrazol-4-yl)benzenesulfonamide, 2-oxopentadecyl-CoA, CHLORIDE ION, ... | | Authors: | Wright, M.H, Clough, B, Rackham, M.D, Brannigan, J.A, Grainger, M, Bottrill, A.R, Heal, W.P, Broncel, M, Serwa, R.A, Mann, D, Leatherbarrow, R.J, Wilkinson, A.J, Holder, A.A, Tate, E.W. | | Deposit date: | 2012-10-13 | | Release date: | 2014-01-15 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Validation of N-Myristoyltransferase as an Antimalarial Drug Target Using an Integrated Chemical Biology Approach.
Nat.Chem., 6, 2014
|
|
2YNE
 
 | | Plasmodium vivax N-myristoyltransferase in complex with a benzothiophene inhibitor | | Descriptor: | 2-(3-piperidin-4-yloxy-1-benzothiophen-2-yl)-5-[(1,3,5-trimethylpyrazol-4-yl)methyl]-1,3,4-oxadiazole, 2-oxopentadecyl-CoA, CHLORIDE ION, ... | | Authors: | Wright, M.H, Clough, B, Rackham, M.D, Brannigan, J.A, Grainger, M, Bottrill, A.R, Heal, W.P, Broncel, M, Serwa, R.A, Mann, D, Leatherbarrow, R.J, Wilkinson, A.J, Holder, A.A, Tate, E.W. | | Deposit date: | 2012-10-13 | | Release date: | 2014-01-15 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Validation of N-Myristoyltransferase as an Antimalarial Drug Target Using an Integrated Chemical Biology Approach.
Nat.Chem., 6, 2014
|
|
2YAL
 
 | | SinR, Master Regulator of biofilm formation in Bacillus subtilis | | Descriptor: | HTH-TYPE TRANSCRIPTIONAL REGULATOR SINR, NICKEL (II) ION | | Authors: | Colledge, V.L, Fogg, M.J, Levdikov, V.M, Leech, A, Dodson, E.J, Wilkinson, A.J. | | Deposit date: | 2011-02-23 | | Release date: | 2011-06-08 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Structure and Organisation of Sinr, the Master Regulator of Biofilm Formation in Bacillus Subtilis.
J.Mol.Biol., 411, 2011
|
|
2YNC
 
 | | Plasmodium vivax N-myristoyltransferase in complex with YnC12-CoA thioester. | | Descriptor: | CHLORIDE ION, DIMETHYL SULFOXIDE, GLYCYLPEPTIDE N-TETRADECANOYLTRANSFERASE, ... | | Authors: | Wright, M.H, Clough, B, Rackham, M.D, Brannigan, J.A, Grainger, M, Bottrill, A.R, Heal, W.P, Broncel, M, Serwa, R.A, Mann, D, Leatherbarrow, R.J, Wilkinson, A.J, Holder, A.A, Tate, E.W. | | Deposit date: | 2012-10-13 | | Release date: | 2014-01-15 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Validation of N-Myristoyltransferase as an Antimalarial Drug Target Using an Integrated Chemical Biology Approach.
Nat.Chem., 6, 2014
|
|
1H4Z
 
 | | Structure of the Anti-Sigma Factor Antagonist SpoIIAA in its Unphosphorylated Form | | Descriptor: | ANTI-SIGMA F FACTOR ANTAGONIST | | Authors: | Seavers, P.R, Lewis, R.J, Brannigan, J.A, Verschueren, K.H.G, Murshudov, G.N, Wilkinson, A.J. | | Deposit date: | 2001-05-16 | | Release date: | 2001-07-06 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.74 Å) | | Cite: | Structure of the Bacillus Cell Fate Determinant Spoiiaa in Phosphorylated and Unphosphorylated Forms
Structure, 9, 2001
|
|
1T9H
 
 | | The crystal structure of YloQ, a circularly permuted GTPase. | | Descriptor: | ACETATE ION, CALCIUM ION, Probable GTPase engC, ... | | Authors: | Levdikov, V.M, Blagova, E.V, Brannigan, J.A, Cladiere, L, Antson, A.A, Isupov, M.N, Seror, S.J, Wilkinson, A.J. | | Deposit date: | 2004-05-17 | | Release date: | 2004-11-02 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The Crystal Structure of YloQ, a Circularly Permuted GTPase Essential for Bacillus Subtilis Viability.
J.Mol.Biol., 340, 2004
|
|
1XOC
 
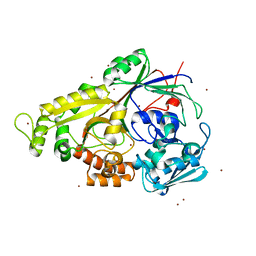 | | The structure of the oligopeptide-binding protein, AppA, from Bacillus subtilis in complex with a nonapeptide. | | Descriptor: | Nonapeptide VDSKNTSSW, Oligopeptide-binding protein appA, ZINC ION | | Authors: | Levdikov, V.M, Blagova, E.V, Brannigan, J.A, Wright, L, Vagin, A.A, Wilkinson, A.J. | | Deposit date: | 2004-10-06 | | Release date: | 2005-01-25 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | The structure of the oligopeptide-binding protein, AppA, from Bacillus subtilis in complex with a nonapeptide.
J.Mol.Biol., 345, 2005
|
|
1OLA
 
 | |
1PMB
 
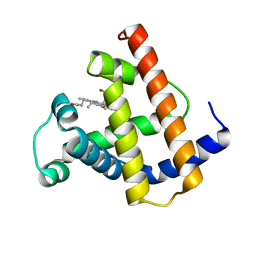 | | THE DETERMINATION OF THE CRYSTAL STRUCTURE OF RECOMBINANT PIG MYOGLOBIN BY MOLECULAR REPLACEMENT AND ITS REFINEMENT | | Descriptor: | MYOGLOBIN, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Smerdon, S.J, Oldfield, T.J, Dodson, E.J, Dodson, G.G, Hubbard, R.E, Wilkinson, A.J. | | Deposit date: | 1989-11-27 | | Release date: | 1990-01-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Determination of the crystal structure of recombinant pig myoglobin by molecular replacement and its refinement.
Acta Crystallogr.,Sect.B, 46, 1990
|
|
1DZ3
 
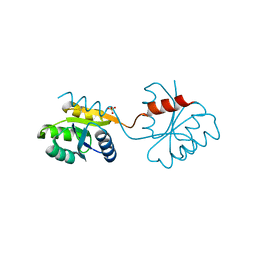 | | DOMAIN-SWAPPING IN THE SPORULATION RESPONSE REGULATOR SPO0A | | Descriptor: | SULFATE ION, Stage 0 sporulation protein A | | Authors: | Lewis, R.J, Brannigan, J.A, Muchova, K, Leonard, G, Barak, I, Wilkinson, A.J. | | Deposit date: | 2000-02-15 | | Release date: | 2000-04-10 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Domain swapping in the sporulation response regulator Spo0A.
J. Mol. Biol., 297, 2000
|
|
1H4X
 
 | | Structure of the Bacillus Cell Fate Determinant SpoIIAA in the Phosphorylated Form | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ANTI-SIGMA F FACTOR ANTAGONIST | | Authors: | Seavers, P.R, Lewis, R.J, Brannigan, J.A, Verschueren, K.H.G, Murshudov, G.N, Wilkinson, A.J. | | Deposit date: | 2001-05-15 | | Release date: | 2001-07-06 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.16 Å) | | Cite: | Structure of the Bacillus Cell Fate Determinant Spoiiaa in Phosphorylated and Unphosphorylated Forms
Structure, 9, 2001
|
|
1H4Y
 
 | | Structure of the Anti-Sigma Factor Antagonist SpoIIAA in its Unphosphorylated Form | | Descriptor: | ANTI-SIGMA F FACTOR ANTAGONIST | | Authors: | Seavers, P.R, Lewis, R.J, Brannigan, J.A, Verschueren, K.H.G, Murshudov, G.N, Wilkinson, A.J. | | Deposit date: | 2001-05-16 | | Release date: | 2001-07-06 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Structure of the Bacillus Cell Fate Determinant Spoiiaa in Phosphorylated and Unphosphorylated Forms
Structure, 9, 2001
|
|
2FYI
 
 | | Crystal Structure of the Cofactor-Binding Domain of the Cbl Transcriptional Regulator | | Descriptor: | HTH-type transcriptional regulator cbl | | Authors: | Stec, E, Neumann, P, Wilkinson, A.J, Brzozowski, A.M, Bujacz, G.D. | | Deposit date: | 2006-02-08 | | Release date: | 2006-02-21 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural Basis of the Sulphate Starvation Response in E. coli: Crystal Structure and Mutational Analysis of the Cofactor-binding Domain of the Cbl Transcriptional Regulator.
J.Mol.Biol., 364, 2006
|
|
6T9M
 
 | | Crystal structure of the Chitinase Domain of the Spore Coat Protein CotE from Clostridium difficile | | Descriptor: | DI(HYDROXYETHYL)ETHER, PENTAETHYLENE GLYCOL, Peptide in active site, ... | | Authors: | Whittingham, J.L, Dodson, E.J, Wilkinson, A.J. | | Deposit date: | 2019-10-28 | | Release date: | 2020-07-22 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Crystal structures of the GH18 domain of the bifunctional peroxiredoxin-chitinase CotE from Clostridium difficile.
Acta Crystallogr.,Sect.F, 76, 2020
|
|
5DOL
 
 | | Crystal structure of YabA amino-terminal domain from Bacillus subtilis | | Descriptor: | Initiation-control protein YabA | | Authors: | Cherrier, M.V, Bazin, A, Jameson, K.H, Wilkinson, A.J, Noirot-Gros, M.F, Terradot, L. | | Deposit date: | 2015-09-11 | | Release date: | 2016-01-20 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Tetramerization and interdomain flexibility of the replication initiation controller YabA enables simultaneous binding to multiple partners.
Nucleic Acids Res., 44, 2016
|
|
6TSB
 
 | |
5G21
 
 | | Leishmania major N-myristoyltransferase in complex with a quinoline inhibitor (compound 26). | | Descriptor: | ETHYL 4-[(2-CYANOETHYL)SULFANYL]-6-{[6-(PIPERAZIN-1-YL), GLYCYLPEPTIDE N-TETRADECANOYLTRANSFERASE, MAGNESIUM ION, ... | | Authors: | Goncalves, V, Brannigan, J.A, Laporte, A, Bell, A.S, Roberts, S.M, Wilkinson, A.J, Leatherbarrow, R.J, Tate, E.W. | | Deposit date: | 2016-04-06 | | Release date: | 2017-02-15 | | Last modified: | 2017-06-28 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure-guided optimization of quinoline inhibitors of Plasmodium N-myristoyltransferase.
Medchemcomm, 8, 2017
|
|
