8OQP
 
 | |
8OPV
 
 | |
8OQM
 
 | |
8OQU
 
 | |
4ZDB
 
 | | Yeast enoyl-CoA isomerase (ScECI2) complexed with acetoacetyl-CoA | | Descriptor: | 3,2-trans-enoyl-CoA isomerase, ACETOACETYL-COENZYME A, GLYCEROL, ... | | Authors: | Onwukwe, G.U, Koski, M.K, Wierenga, R.K. | | Deposit date: | 2015-04-17 | | Release date: | 2015-11-11 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Structures of yeast peroxisomal Delta (3), Delta (2)-enoyl-CoA isomerase complexed with acyl-CoA substrate analogues: the importance of hydrogen-bond networks for the reactivity of the catalytic base and the oxyanion hole.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4ZDF
 
 | | Crystal structure of yeast enoyl-CoA isomerase helix-10 deletion (ScECI2-H10) mutant | | Descriptor: | 3,2-trans-enoyl-CoA isomerase, GLYCEROL | | Authors: | Onwukwe, G.U, Koski, M.K, Wierenga, R.K. | | Deposit date: | 2015-04-17 | | Release date: | 2015-11-11 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Structures of yeast peroxisomal Delta (3), Delta (2)-enoyl-CoA isomerase complexed with acyl-CoA substrate analogues: the importance of hydrogen-bond networks for the reactivity of the catalytic base and the oxyanion hole.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4ZDE
 
 | | Crystal structure of yeast D3,D2-enoyl-CoA isomerase F268A mutant | | Descriptor: | 3,2-trans-enoyl-CoA isomerase, GLYCEROL, SULFATE ION | | Authors: | Onwukwe, G.U, Koski, M.K, Wierenga, R.K. | | Deposit date: | 2015-04-17 | | Release date: | 2015-11-11 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structures of yeast peroxisomal Delta (3), Delta (2)-enoyl-CoA isomerase complexed with acyl-CoA substrate analogues: the importance of hydrogen-bond networks for the reactivity of the catalytic base and the oxyanion hole.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4ZDC
 
 | | Yeast enoyl-CoA isomerase complexed with octanoyl-CoA | | Descriptor: | 3,2-trans-enoyl-CoA isomerase, GLYCEROL, OCTANOYL-COENZYME A, ... | | Authors: | Onwukwe, G.U, Koski, M.K, Wierenga, R.K. | | Deposit date: | 2015-04-17 | | Release date: | 2015-11-11 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | Structures of yeast peroxisomal Delta (3), Delta (2)-enoyl-CoA isomerase complexed with acyl-CoA substrate analogues: the importance of hydrogen-bond networks for the reactivity of the catalytic base and the oxyanion hole.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4ZDD
 
 | | Structure of yeast D3,D2-enoyl-CoA isomerase bound to sulphate ion | | Descriptor: | 3,2-trans-enoyl-CoA isomerase, SULFATE ION | | Authors: | Onwukwe, G.U, Koski, M.K, Wierenga, R.K. | | Deposit date: | 2015-04-17 | | Release date: | 2015-11-11 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structures of yeast peroxisomal Delta (3), Delta (2)-enoyl-CoA isomerase complexed with acyl-CoA substrate analogues: the importance of hydrogen-bond networks for the reactivity of the catalytic base and the oxyanion hole.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
3ZWC
 
 | | CRYSTAL STRUCTURE OF RAT PEROXISOMAL MULTIFUNCTIONAL ENZYME TYPE 1 (RPMFE1) COMPLEXED WITH 3S-HYDROXY-DECANOYL-COA | | Descriptor: | (S)-3-HYDROXYDECANOYL-COA, GLYCEROL, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Kasaragod, P, Schmitz, W, Hiltunen, J.K, Wierenga, R.K. | | Deposit date: | 2011-07-28 | | Release date: | 2012-08-08 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The Isomerase and Hydratase Reaction Mechanism of the Crotonase Active Site of the Multifunctional Enzyme (Type-1), as Deduced from Structures of Complexes with 3S-Hydroxy- Acyl-Coa.
FEBS J., 280, 2013
|
|
3ZW8
 
 | | Crystal Structure Of Rat Peroxisomal Multifunctional Enzyme Type 1 (rpMFE1) In Apo Form | | Descriptor: | GLYCEROL, PEROXISOMAL BIFUNCTIONAL ENZYME, SULFATE ION | | Authors: | Kasaragod, P, Schmitz, W, Hiltunen, J.K, Wierenga, R.K. | | Deposit date: | 2011-07-28 | | Release date: | 2012-08-08 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The Isomerase and Hydratase Reaction Mechanism of the Crotonase Active Site of the Multifunctional Enzyme (Type-1), as Deduced from Structures of Complexes with 3S-Hydroxy- Acyl-Coa.
FEBS J., 280, 2013
|
|
3ZW9
 
 | | CRYSTAL STRUCTURE OF RAT PEROXISOMAL MULTIFUNCTIONAL ENZYME TYPE 1 (RPMFE1) COMPLEXED WITH (2S,3S)-3-HYDROXY-2-METHYLBUTANOYL-COA | | Descriptor: | (2S,3S)-3-HYDROXY-2-METHYLBUTANOYL-COA, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, PEROXISOMAL BIFUNCTIONAL ENZYME, ... | | Authors: | Kasaragod, P, Schmitz, W, Hiltunen, J.K, Wierenga, R.K. | | Deposit date: | 2011-07-28 | | Release date: | 2012-08-08 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | The Isomerase and Hydratase Reaction Mechanism of the Crotonase Active Site of the Multifunctional Enzyme (Type-1), as Deduced from Structures of Complexes with 3S-Hydroxy- Acyl-Coa.
FEBS J., 280, 2013
|
|
1M4S
 
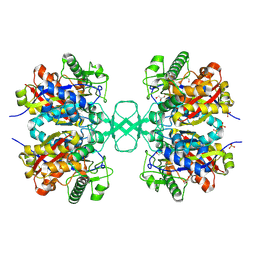 | | Biosynthetic thiolase, Cys89 acetylated, unliganded form | | Descriptor: | Acetyl-CoA acetyltransferase, GLYCEROL, SULFATE ION | | Authors: | Kursula, P, Ojala, J, Lambeir, A.-M, Wierenga, R.K. | | Deposit date: | 2002-07-03 | | Release date: | 2002-11-29 | | Last modified: | 2018-03-07 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | The catalytic cycle of biosynthetic thiolase: A conformational
journey of an acetyl group through four binding modes and two oxyanion holes
Biochemistry, 41, 2002
|
|
3ZWB
 
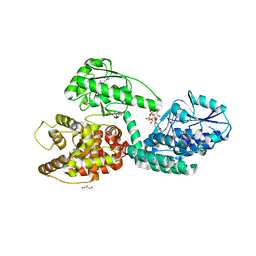 | | CRYSTAL STRUCTURE OF RAT PEROXISOMAL MULTIFUNCTIONAL ENZYME TYPE 1 (RPMFE1) COMPLEXED WITH 2TRANS-HEXENOYL-COA | | Descriptor: | (2E)-Hexenoyl-CoA, GLYCEROL, PEROXISOMAL BIFUNCTIONAL ENZYME, ... | | Authors: | Kasaragod, P, Schmitz, W, Hiltunen, J.K, Wierenga, R.K. | | Deposit date: | 2011-07-28 | | Release date: | 2012-08-08 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | The Isomerase and Hydratase Reaction Mechanism of the Crotonase Active Site of the Multifunctional Enzyme (Type-1), as Deduced from Structures of Complexes with 3S-Hydroxy- Acyl-Coa.
FEBS J., 280, 2013
|
|
4B3H
 
 | |
1M1T
 
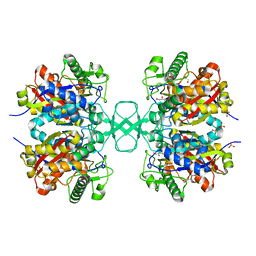 | | Biosynthetic thiolase, Q64A mutant | | Descriptor: | Acetyl-CoA acetyltransferase, GLYCEROL, SULFATE ION | | Authors: | Kursula, P, Ojala, J, Lambeir, A.-M, Wierenga, R.K. | | Deposit date: | 2002-06-20 | | Release date: | 2002-11-29 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | The catalytic cycle of biosynthetic thiolase: A conformational
journey of an acetyl group through four binding modes and two oxyanion holes
Biochemistry, 41, 2002
|
|
1M3Z
 
 | | Biosynthetic thiolase, C89A mutant, complexed with acetyl coenzyme A | | Descriptor: | ACETYL COENZYME *A, Acetyl-CoA acetyltransferase, SULFATE ION | | Authors: | Kursula, P, Ojala, J, Lambeir, A.-M, Wierenga, R.K. | | Deposit date: | 2002-07-03 | | Release date: | 2002-11-29 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | The catalytic cycle of biosynthetic thiolase: A conformational
journey of an acetyl group through four binding modes and two oxyanion holes
Biochemistry, 41, 2002
|
|
1M4T
 
 | | Biosynthetic thiolase, Cys89 butyrylated | | Descriptor: | Acetyl-CoA acetyltransferase, GLYCEROL, SULFATE ION | | Authors: | Kursula, P, Ojala, J, Lambeir, A.-M, Wierenga, R.K. | | Deposit date: | 2002-07-03 | | Release date: | 2002-11-29 | | Last modified: | 2018-03-07 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | The catalytic cycle of biosynthetic thiolase: A conformational
journey of an acetyl group through four binding modes and two oxyanion holes
Biochemistry, 41, 2002
|
|
1M3K
 
 | | biosynthetic thiolase, inactive C89A mutant | | Descriptor: | Acetyl-CoA acetyltransferase, GLYCEROL, SULFATE ION | | Authors: | Kursula, P, Ojala, J, Lambeir, A.-M, Wierenga, R.K. | | Deposit date: | 2002-06-28 | | Release date: | 2002-11-29 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The catalytic cycle of biosynthetic thiolase: A conformational
journey of an acetyl group through four binding modes and two oxyanion holes
Biochemistry, 41, 2002
|
|
1NL7
 
 | |
5AB5
 
 | |
2YIM
 
 | | The enolisation chemistry of a thioester-dependent racemase: the 1.4 A crystal structure of a complex with a planar reaction intermediate analogue | | Descriptor: | 2-METHYLACETOACETYL COA, GLYCEROL, PHOSPHATE ION, ... | | Authors: | Sharma, S, Bhaumik, P, Venkatesan, R, Hiltunen, J.K, Conzelmann, E, Juffer, A.H, Wierenga, R.K. | | Deposit date: | 2011-05-16 | | Release date: | 2012-03-28 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.41 Å) | | Cite: | The Enolization Chemistry of a Thioester-Dependent Racemase: The 1.4 A Crystal Structure of a Reaction Intermediate Complex Characterized by Detailed Qm/Mm Calculations.
J Phys Chem B, 116, 2012
|
|
1M1O
 
 | | Crystal structure of biosynthetic thiolase, C89A mutant, complexed with acetoacetyl-CoA | | Descriptor: | ACETOACETYL-COENZYME A, Acetyl-CoA acetyltransferase, SULFATE ION | | Authors: | Kursula, P, Ojala, J, Lambeir, A.-M, Wierenga, R.K. | | Deposit date: | 2002-06-20 | | Release date: | 2002-11-29 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | The catalytic cycle of biosynthetic thiolase: A conformational journey of an acetyl group through four binding modes and two oxyanion holes
Biochemistry, 41, 2002
|
|
8PF8
 
 | | Structure of Mycobacterium tuberculosis beta-oxidation trifunctional enzyme in complex with Fragment-M-72 | | Descriptor: | (2~{R})-3-bis[2-methyl-5-(trifluoromethyl)pyrazol-3-yl]boranyloxypropane-1,2-diol, 3-hydroxyacyl-CoA dehydrogenase, GLYCEROL, ... | | Authors: | Dalwani, S, Wierenga, R.K, Venkatesan, R. | | Deposit date: | 2023-06-15 | | Release date: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Crystallographic fragment binding studies of the Mycobacterium tuberculosis trifunctional enzyme suggest binding pockets for the tails of the acyl-CoA substrates at its active sites and a potential substrate channeling path between them
Biorxiv, 2024
|
|
5AB4
 
 | |
