2QB7
 
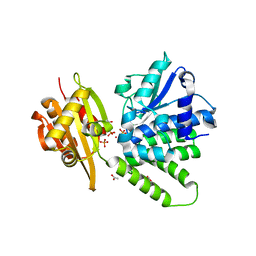 | | Saccharomyces cerevisiae cytosolic exopolyphosphatase, phosphate complex | | 分子名称: | 1,2-ETHANEDIOL, ACETATE ION, COBALT (II) ION, ... | | 著者 | White, S.A, Ugochukwu, E. | | 登録日 | 2007-06-16 | | 公開日 | 2007-12-11 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | The crystal structure of the cytosolic exopolyphosphatase from Saccharomyces cerevisiae reveals the basis for substrate specificity.
J.Mol.Biol., 371, 2007
|
|
7Q0O
 
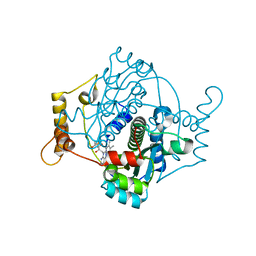 | | E. coli NfsA | | 分子名称: | FLAVIN MONONUCLEOTIDE, Oxygen-insensitive NADPH nitroreductase | | 著者 | White, S.A, Grainger, A, Parr, R, Day, M.A, Jarrom, D, Graziano, A, Searle, P.F, Hyde, E.I. | | 登録日 | 2021-10-15 | | 公開日 | 2022-06-22 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (0.96 Å) | | 主引用文献 | The 3D-structure, kinetics and dynamics of the E. coli nitroreductase NfsA with NADP + provide glimpses of its catalytic mechanism.
Febs Lett., 596, 2022
|
|
7Z0W
 
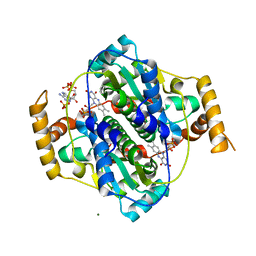 | | E. coli NfsA bound to NADP+ | | 分子名称: | 2'-MONOPHOSPHOADENOSINE-5'-DIPHOSPHATE, FLAVIN MONONUCLEOTIDE, MAGNESIUM ION, ... | | 著者 | White, S.A, Grainger, A, Parr, R, Day, M.A, Jarrom, D, Graziano, A, Searle, P.F, Hyde, E.I. | | 登録日 | 2022-02-23 | | 公開日 | 2022-07-20 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (2.06 Å) | | 主引用文献 | The 3D-structure, kinetics and dynamics of the E. coli nitroreductase NfsA with NADP + provide glimpses of its catalytic mechanism.
Febs Lett., 596, 2022
|
|
7P3H
 
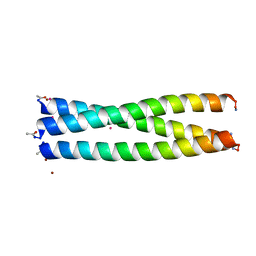 | |
8CJ0
 
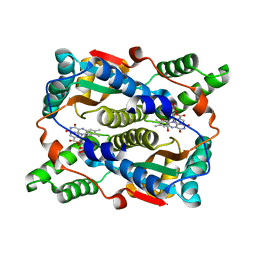 | | E. coli NfsB-T41Q/N71S/F124T/M127V mutant bound to nicotinate | | 分子名称: | 1,2-ETHANEDIOL, FLAVIN MONONUCLEOTIDE, NICOTINIC ACID, ... | | 著者 | White, S.A, Hyde, E.I, Day, M.A. | | 登録日 | 2023-02-11 | | 公開日 | 2023-04-19 | | 最終更新日 | 2024-06-19 | | 実験手法 | X-RAY DIFFRACTION (1.99 Å) | | 主引用文献 | Structure and Dynamics of Three Escherichia coli NfsB Nitro-Reductase Mutants Selected for Enhanced Activity with the Cancer Prodrug CB1954.
Int J Mol Sci, 24, 2023
|
|
8C5E
 
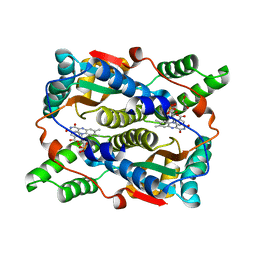 | | E. coli NfsB-T41Q/N71S/F124T mutant bound to nicotinic acid | | 分子名称: | 1,2-ETHANEDIOL, FLAVIN MONONUCLEOTIDE, NICOTINIC ACID, ... | | 著者 | White, S.A, Hyde, E.I, Day, M.A. | | 登録日 | 2023-01-06 | | 公開日 | 2023-04-19 | | 最終更新日 | 2024-06-19 | | 実験手法 | X-RAY DIFFRACTION (1.65 Å) | | 主引用文献 | Structure and Dynamics of Three Escherichia coli NfsB Nitro-Reductase Mutants Selected for Enhanced Activity with the Cancer Prodrug CB1954.
Int J Mol Sci, 24, 2023
|
|
8C5F
 
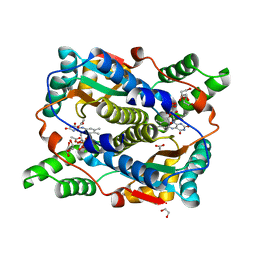 | | E. coli NfsB-T41Q/N71S/F124T mutant bound to acetate | | 分子名称: | 1,2-ETHANEDIOL, ACETATE ION, FLAVIN MONONUCLEOTIDE, ... | | 著者 | White, S.A, Hyde, E.I, Day, M.A. | | 登録日 | 2023-01-07 | | 公開日 | 2023-04-19 | | 最終更新日 | 2024-06-19 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Structure and Dynamics of Three Escherichia coli NfsB Nitro-Reductase Mutants Selected for Enhanced Activity with the Cancer Prodrug CB1954.
Int J Mol Sci, 24, 2023
|
|
3H7U
 
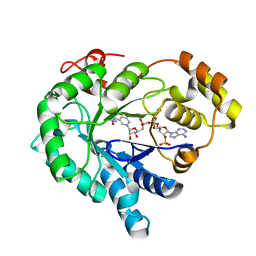 | | Crystal structure of the plant stress-response enzyme AKR4C9 | | 分子名称: | ACETATE ION, Aldo-keto reductase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | 著者 | White, S.A, Simpson, P.J, Ride, J.P. | | 登録日 | 2009-04-28 | | 公開日 | 2009-08-04 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (1.25 Å) | | 主引用文献 | Characterization of two novel aldo-keto reductases from Arabidopsis: expression patterns, broad substrate specificity, and an open active-site structure suggest a role in toxicant metabolism following stress.
J.Mol.Biol., 392, 2009
|
|
3H7R
 
 | | Crystal structure of the plant stress-response enzyme AKR4C8 | | 分子名称: | 1,2-ETHANEDIOL, ACETATE ION, Aldo-keto reductase, ... | | 著者 | White, S.A, Simpson, P.J, Ride, J.P. | | 登録日 | 2009-04-28 | | 公開日 | 2009-08-04 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (1.4 Å) | | 主引用文献 | Characterization of two novel aldo-keto reductases from Arabidopsis: expression patterns, broad substrate specificity, and an open active-site structure suggest a role in toxicant metabolism following stress.
J.Mol.Biol., 392, 2009
|
|
5CLV
 
 | | Crystal Structure of KorA-operator DNA complex (KorA-OA) | | 分子名称: | 5'-D(CP*CP*AP*AP*GP*TP*TP*TP*AP*GP*CP*TP*AP*AP*AP*CP*TP*TP*GP*GP*)-3', TrfB transcriptional repressor protein | | 著者 | White, S.A, Hyde, E.I, Rajasekar, K.V. | | 登録日 | 2015-07-16 | | 公開日 | 2016-04-06 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Flexibility of KorA, a plasmid-encoded, global transcription regulator, in the presence and the absence of its operator.
Nucleic Acids Res., 44, 2016
|
|
5CKT
 
 | | Crystal Structure of KorA, a plasmid-encoded, global transcription regulator | | 分子名称: | ACETATE ION, TrfB transcriptional repressor protein | | 著者 | White, S.A, Hyde, E.I, Lovering, A.L. | | 登録日 | 2015-07-15 | | 公開日 | 2016-04-06 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Flexibility of KorA, a plasmid-encoded, global transcription regulator, in the presence and the absence of its operator.
Nucleic Acids Res., 44, 2016
|
|
5CM3
 
 | | Crystal Structure of KorA, a plasmid-encoded, global transcription regulator | | 分子名称: | 5'-D(CP*CP*AP*AP*GP*TP*TP*TP*AP*GP*CP*TP*AP*AP*AP*CP*TP*TP*GP*GP*)-3', TrfB transcriptional repressor protein | | 著者 | White, S.A, Hyde, E.I, Rajasekar, K.V. | | 登録日 | 2015-07-16 | | 公開日 | 2016-04-06 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (2.302 Å) | | 主引用文献 | Flexibility of KorA, a plasmid-encoded, global transcription regulator, in the presence and the absence of its operator.
Nucleic Acids Res., 44, 2016
|
|
3FIQ
 
 | | Odorant Binding Protein OBP1 | | 分子名称: | 1,2-ETHANEDIOL, Odorant-binding protein 1F | | 著者 | White, S.A. | | 登録日 | 2008-12-12 | | 公開日 | 2009-05-19 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (1.599 Å) | | 主引用文献 | Structure of rat odorant-binding protein OBP1 at 1.6 A resolution
Acta Crystallogr.,Sect.D, 65, 2009
|
|
3O9O
 
 | |
2QB8
 
 | |
2QB6
 
 | | Saccharomyces cerevisiae cytosolic exopolyphosphatase, sulfate complex | | 分子名称: | 1,2-ETHANEDIOL, ACETATE ION, Exopolyphosphatase, ... | | 著者 | White, S.A, Ugochukwu, E. | | 登録日 | 2007-06-16 | | 公開日 | 2007-12-11 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | The crystal structure of the cytosolic exopolyphosphatase from Saccharomyces cerevisiae reveals the basis for substrate specificity.
J.Mol.Biol., 371, 2007
|
|
6GOX
 
 | | SecA | | 分子名称: | Protein translocase subunit SecA | | 著者 | White, S.A, Huber, D. | | 登録日 | 2018-06-04 | | 公開日 | 2019-06-19 | | 最終更新日 | 2024-05-15 | | 実験手法 | X-RAY DIFFRACTION (3.5 Å) | | 主引用文献 | The C-terminal tail of the bacterial translocation ATPase SecA modulates its activity.
Elife, 8, 2019
|
|
1DJL
 
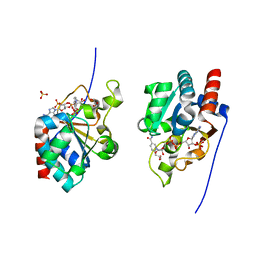 | | THE CRYSTAL STRUCTURE OF HUMAN TRANSHYDROGENASE DOMAIN III WITH BOUND NADP | | 分子名称: | GLYCEROL, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, SULFATE ION, ... | | 著者 | White, S.A, Peak, S.J, Cotton, N.P, Jackson, J.B. | | 登録日 | 1999-12-03 | | 公開日 | 2000-12-06 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | The high-resolution structure of the NADP(H)-binding component (dIII) of proton-translocating transhydrogenase from human heart mitochondria.
Structure Fold.Des., 8, 2000
|
|
8OG3
 
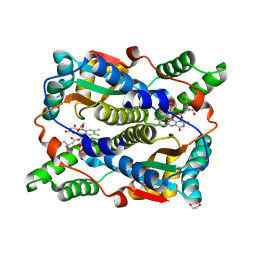 | | E. coli NfsB triple mutant T41L/N71S/F124T bound to citrate | | 分子名称: | 1,2-ETHANEDIOL, CITRIC ACID, FLAVIN MONONUCLEOTIDE, ... | | 著者 | Day, M.A, White, S.A, Hyde, E.I, Searle, P.F. | | 登録日 | 2023-03-17 | | 公開日 | 2023-04-19 | | 最終更新日 | 2024-06-19 | | 実験手法 | X-RAY DIFFRACTION (2.09 Å) | | 主引用文献 | Structure and Dynamics of Three Escherichia coli NfsB Nitro-Reductase Mutants Selected for Enhanced Activity with the Cancer Prodrug CB1954.
Int J Mol Sci, 24, 2023
|
|
8QES
 
 | |
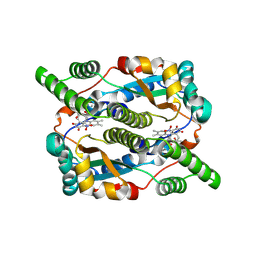
8QPO
 
 | |
8AJX
 
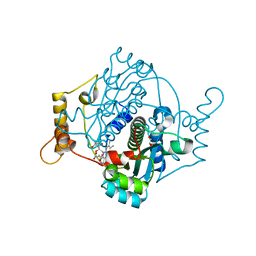 | | E. coli NfsA with Fumarate | | 分子名称: | 1,2-ETHANEDIOL, FLAVIN MONONUCLEOTIDE, FUMARIC ACID, ... | | 著者 | Day, M.A, Jarrom, D, White, S.A, Hyde, E.I. | | 登録日 | 2022-07-28 | | 公開日 | 2023-01-18 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (1.25 Å) | | 主引用文献 | Oxygen-insensitive nitroreductase E. coli NfsA, but not NfsB, is inhibited by fumarate.
Proteins, 91, 2023
|
|
8C5P
 
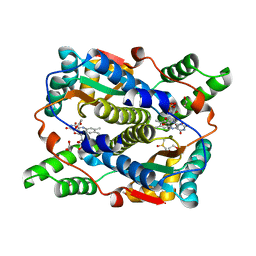 | | E. coli NfsB mutant N71S T41L with acetate | | 分子名称: | 1,2-ETHANEDIOL, ACETATE ION, DIMETHYL SULFOXIDE, ... | | 著者 | Day, M.A, White, S.A, Hyde, E.I. | | 登録日 | 2023-01-10 | | 公開日 | 2023-04-19 | | 最終更新日 | 2024-06-19 | | 実験手法 | X-RAY DIFFRACTION (1.69 Å) | | 主引用文献 | Structure and Dynamics of Three Escherichia coli NfsB Nitro-Reductase Mutants Selected for Enhanced Activity with the Cancer Prodrug CB1954.
Int J Mol Sci, 24, 2023
|
|
8CCV
 
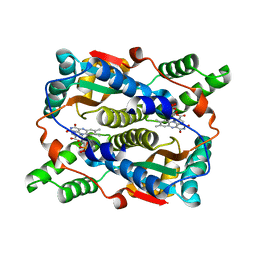 | | E. coli NfsB mutant T41LN71S with nicotinate | | 分子名称: | FLAVIN MONONUCLEOTIDE, NICOTINIC ACID, Oxygen-insensitive NAD(P)H nitroreductase | | 著者 | Day, M.A, White, S.A, Hyde, E.I, Searle, P.F. | | 登録日 | 2023-01-27 | | 公開日 | 2023-04-19 | | 最終更新日 | 2024-06-19 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Structure and Dynamics of Three Escherichia coli NfsB Nitro-Reductase Mutants Selected for Enhanced Activity with the Cancer Prodrug CB1954.
Int J Mol Sci, 24, 2023
|
|
5SWC
 
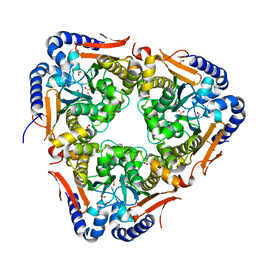 | | The structure of the beta-carbonic anhydrase CcaA | | 分子名称: | CHLORIDE ION, Carbonic anhydrase, FORMIC ACID, ... | | 著者 | Kimber, M.S, McGurn, L, White, S.A. | | 登録日 | 2016-08-08 | | 公開日 | 2016-10-26 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (1.45 Å) | | 主引用文献 | The structure, kinetics and interactions of the beta-carboxysomal beta-carbonic anhydrase, CcaA.
Biochem. J., 473, 2016
|
|
