1WHI
 
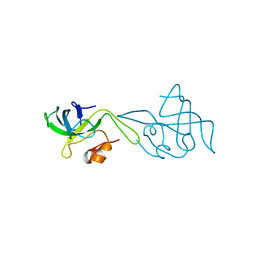 | | RIBOSOMAL PROTEIN L14 | | Descriptor: | RIBOSOMAL PROTEIN L14 | | Authors: | Davies, C, White, S.W, Ramakrishnan, V. | | Deposit date: | 1996-01-10 | | Release date: | 1996-08-17 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The crystal structure of ribosomal protein L14 reveals an important organizational component of the translational apparatus.
Structure, 4, 1996
|
|
1G5X
 
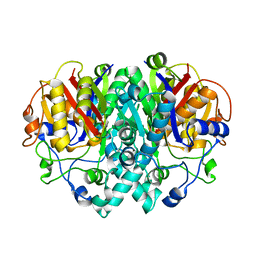 | | The Structure of Beta-Ketoacyl-[Acyl Carrier Protein] Synthase I | | Descriptor: | BETA-KETOACYL ACYL CARRIER PROTEIN SYNTHASE I | | Authors: | Zhang, Y.M, Rao, M.S, Heath, R.J, Price, A.C, Olson, A.J, Rock, C.O, White, S.W. | | Deposit date: | 2000-11-02 | | Release date: | 2000-11-15 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Identification and analysis of the acyl carrier protein (ACP) docking site on beta-ketoacyl-ACP synthase III.
J.Biol.Chem., 276, 2001
|
|
7NB9
 
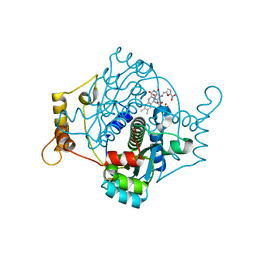 | | E. coli NfsA with nitrofurantoin | | Descriptor: | 1-[(~{E})-(5-nitrofuran-2-yl)methylideneamino]imidazolidine-2,4-dione, DIMETHYL SULFOXIDE, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Day, M.D, Jarrom, D, Grainger, A.I, Parr, R.J, Hyde, E.I, White, S.A. | | Deposit date: | 2021-01-25 | | Release date: | 2021-07-21 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.09 Å) | | Cite: | The structures of E. coli NfsA bound to the antibiotic nitrofurantoin; to 1,4-benzoquinone and to FMN.
Biochem.J., 478, 2021
|
|
7NNX
 
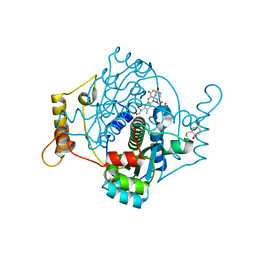 | | E. coli NfsA with 1,4-benzoquinone | | Descriptor: | 1,2-ETHANEDIOL, 1,4-benzoquinone, CHLORIDE ION, ... | | Authors: | Day, M.D, Jarrom, D, Hyde, E.I, White, S.A. | | Deposit date: | 2021-02-25 | | Release date: | 2021-07-21 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The structures of E. coli NfsA bound to the antibiotic nitrofurantoin; to 1,4-benzoquinone and to FMN.
Biochem.J., 478, 2021
|
|
7NMP
 
 | | E. coli NfsA with hydroquinone | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Day, M.D, Jarrom, D, Parr, R.J, Hyde, E.I, White, S.A. | | Deposit date: | 2021-02-23 | | Release date: | 2021-07-21 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | The structures of E. coli NfsA bound to the antibiotic nitrofurantoin; to 1,4-benzoquinone and to FMN.
Biochem.J., 478, 2021
|
|
1YKI
 
 | | The structure of E. coli nitroreductase bound with the antibiotic nitrofurazone | | Descriptor: | CITRIC ACID, DIMETHYL SULFOXIDE, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Race, P.R, Lovering, A.L, Green, R.M, Ossor, A, White, S.A, Searle, P.F, Wrighton, C.J, Hyde, E.I. | | Deposit date: | 2005-01-18 | | Release date: | 2005-02-08 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural and mechanistic studies of Escherichia coli nitroreductase with the antibiotic nitrofurazone. Reversed binding orientations in different redox states of the enzyme.
J.Biol.Chem., 280, 2005
|
|
1F8G
 
 | | THE X-RAY STRUCTURE OF NICOTINAMIDE NUCLEOTIDE TRANSHYDROGENASE FROM RHODOSPIRILLUM RUBRUM COMPLEXED WITH NAD+ | | Descriptor: | NICOTINAMIDE NUCLEOTIDE TRANSHYDROGENASE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Buckley, P.A, Baz Jackson, J, Schneider, T, White, S.A, Rice, D.W, Baker, P.J. | | Deposit date: | 2000-06-30 | | Release date: | 2001-06-30 | | Last modified: | 2017-09-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Protein-protein recognition, hydride transfer and proton pumping in the transhydrogenase complex.
Structure Fold.Des., 8, 2000
|
|
4YWO
 
 | | Mercuric reductase from Metallosphaera sedula | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, Mercuric reductase | | Authors: | Artz, J.H, Zadvornyy, O.A, White, S, Peters, J.W. | | Deposit date: | 2015-03-20 | | Release date: | 2015-09-16 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Biochemical and Structural Properties of a Thermostable Mercuric Ion Reductase from Metallosphaera sedula.
Front Bioeng Biotechnol, 3, 2015
|
|
7NIY
 
 | | E. coli NfsA with FMN | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Day, M.D, Jarrom, D, Hyde, E.I, White, S.A. | | Deposit date: | 2021-02-14 | | Release date: | 2021-07-21 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.03 Å) | | Cite: | The structures of E. coli NfsA bound to the antibiotic nitrofurantoin; to 1,4-benzoquinone and to FMN.
Biochem.J., 478, 2021
|
|
6W6B
 
 | | The X-ray crystal structure of the C-terminus domain of Staphylococcus aureus Fatty Acid Kinase A (FakA, residues 328-548) protein to 1.40 Angstrom resolution | | Descriptor: | SULFATE ION, SaFakA-Cterminus domain | | Authors: | Cuypers, M.G, Subramanian, C, Rock, C.O, White, S.W. | | Deposit date: | 2020-03-16 | | Release date: | 2021-03-17 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | The X-ray crystal structure of the C-terminus domain of Staphylococcus aureus Fatty Acid Kinase A (FakA, residues 328-548) protein to 1.40 Angstrom resolution
To Be Published
|
|
6W7A
 
 | | The crystal structure of the 2009/H1N1/California PA endonuclease mutant E119D bound to DNA oligomer TAGCAT (uncleaved, 5mM overnight DNA soak) | | Descriptor: | DNA (5'-D(P*TP*AP*GP*CP*AP*T)-3'), GLYCEROL, MANGANESE (II) ION, ... | | Authors: | Cuypers, M.G, Kumar, G, Webb, T, White, S.W. | | Deposit date: | 2020-03-18 | | Release date: | 2021-02-03 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Structural insights into the substrate specificity of the endonuclease activity of the influenza virus cap-snatching mechanism.
Nucleic Acids Res., 49, 2021
|
|
6WHM
 
 | | The crystal structure of the 2009/H1N1/California PA endonuclease wild type bound to DNA oligomer TAGC (cleaved TTAGCATT, 5mM overnight DNA soak) | | Descriptor: | DNA (5'-D(P*TP*AP*GP*C)-3'), GLYCEROL, MAGNESIUM ION, ... | | Authors: | Cuypers, M.G, Kumar, G, Webb, T, White, S.W. | | Deposit date: | 2020-04-08 | | Release date: | 2021-02-03 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Structural insights into the substrate specificity of the endonuclease activity of the influenza virus cap-snatching mechanism.
Nucleic Acids Res., 49, 2021
|
|
6WS3
 
 | | The crystal structure of the 2009/H1N1/California PA endonuclease wild type bound to DNA oligomers TG and AGCA (from cleaved GTGAGCAGTG) | | Descriptor: | DNA (5'-D(P*TP*G)-3'), GLYCEROL, MANGANESE (II) ION, ... | | Authors: | Cuypers, M.G, Kumar, G, Webb, T, White, S.W. | | Deposit date: | 2020-04-30 | | Release date: | 2021-02-03 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural insights into the substrate specificity of the endonuclease activity of the influenza virus cap-snatching mechanism.
Nucleic Acids Res., 49, 2021
|
|
6PC4
 
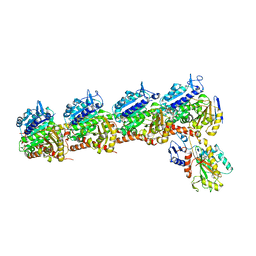 | | Tubulin-RB3_SLD-TTL in complex with compound ABI-274 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Kumar, G, Wang, Y, Li, W, White, S.W. | | Deposit date: | 2019-06-15 | | Release date: | 2020-04-22 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.602 Å) | | Cite: | Structure-Activity Relationship Study of Novel 6-Aryl-2-benzoyl-pyridines as Tubulin Polymerization Inhibitors with Potent Antiproliferative Properties.
J.Med.Chem., 63, 2020
|
|
5KYM
 
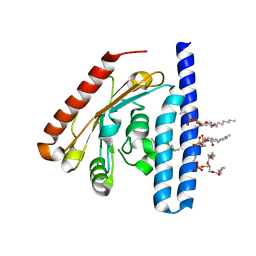 | | Crystal Structure of the 1-acyl-sn-glycerophosphate (LPA) acyltransferase, PlsC, from Thermotoga maritima | | Descriptor: | 1-HEPTADECANOYL-2-TRIDECANOYL-3-GLYCEROL-PHOSPHONYL CHOLINE, 1-acyl-sn-glycerol-3-phosphate acyltransferase, DODECYL-BETA-D-MALTOSIDE, ... | | Authors: | Robertson, R.M, Yao, J, Gajewski, S, Kumar, G, Martin, E.W, Rock, C.O, White, S.W. | | Deposit date: | 2016-07-21 | | Release date: | 2017-07-26 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | A two-helix motif positions the lysophosphatidic acid acyltransferase active site for catalysis within the membrane bilayer.
Nat. Struct. Mol. Biol., 24, 2017
|
|
6NYU
 
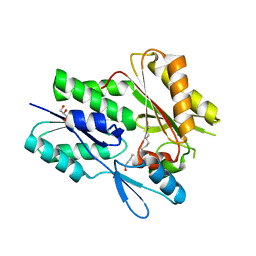 | | The X-ray crystal structure of Staphylococcus aureus Fatty Acid Kinase (Fak) B1 F263T mutant protein to 2.18 Angstrom resolution | | Descriptor: | DegV domain-containing protein, GLYCEROL, MYRISTIC ACID, ... | | Authors: | Cuypers, M.G, Gullett, J.M, Subramanian, C, White, S.W, Rock, C.O. | | Deposit date: | 2019-02-12 | | Release date: | 2020-02-12 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.183 Å) | | Cite: | The X-ray crystal structure of STAPHYLOCOCCUS AUREUS Fatty Acid Kinase (Fak) B1 protein F263T mutant to 2.18 Angstrom resolution
To Be Published
|
|
6NR1
 
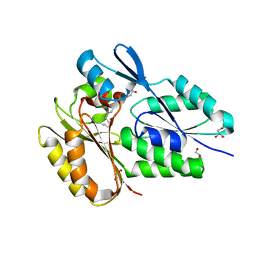 | | The X-ray crystal structure of Streptococcus pneumoniae Fatty Acid Kinase (Fak) B2 protein loaded with Vaccenic acid (C18:1 delta11) to 2.1 Angstrom resolution | | Descriptor: | Fatty Acid Kinase (Fak) B2 protein (SPR1019), GLYCEROL, VACCENIC ACID | | Authors: | Cuypers, M.G, Gullett, J.M, Subramanian, C, White, S.W, Rock, C.O. | | Deposit date: | 2019-01-22 | | Release date: | 2019-08-21 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The X-ray crystal structure of Streptococcus pneumoniae Fatty Acid Kinase (Fak) B2 protein loaded with Vaccenic acid (C18:1 delta11) to 2.1 Angstrom resolution
To Be Published
|
|
4HNJ
 
 | |
5W9G
 
 | | Crystal structure of the influenza virus PA endonuclease in complex with inhibitor 9k (SRI-30023) | | Descriptor: | 2-[(2S)-1-{[(2-chlorophenyl)sulfanyl]acetyl}pyrrolidin-2-yl]-5-hydroxy-6-oxo-N-(2-phenoxyethyl)-1,6-dihydropyrimidine-4 -carboxamide, CHLORIDE ION, MANGANESE (II) ION, ... | | Authors: | Kumar, G, White, S. | | Deposit date: | 2017-06-23 | | Release date: | 2017-12-27 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Protein-Structure Assisted Optimization of 4,5-Dihydroxypyrimidine-6-Carboxamide Inhibitors of Influenza Virus Endonuclease.
Sci Rep, 7, 2017
|
|
5CGV
 
 | | 2009 H1N1 PA endonuclease in complex with L-742,001 | | Descriptor: | (2Z)-4-[1-benzyl-4-(4-chlorobenzyl)piperidin-4-yl]-2-hydroxy-4-oxobut-2-enoic acid, GLYCEROL, MANGANESE (II) ION, ... | | Authors: | Kumar, G, White, S.W. | | Deposit date: | 2015-07-09 | | Release date: | 2016-03-23 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Identification and characterization of influenza variants resistant to a viral endonuclease inhibitor.
Proc. Natl. Acad. Sci. U.S.A., 113, 2016
|
|
1YLU
 
 | | The structure of E. coli nitroreductase with bound acetate, crystal form 2 | | Descriptor: | ACETATE ION, FLAVIN MONONUCLEOTIDE, Oxygen-insensitive NAD(P)H nitroreductase | | Authors: | Race, P.R, Lovering, A.L, Green, R.M, Ossor, A, White, S.A, Searle, P.F, Wrighton, C.J, Hyde, E.I. | | Deposit date: | 2005-01-19 | | Release date: | 2005-02-08 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural and mechanistic studies of Escherichia coli nitroreductase with the antibiotic nitrofurazone. Reversed binding orientations in different redox states of the enzyme.
J.Biol.Chem., 280, 2005
|
|
1YLR
 
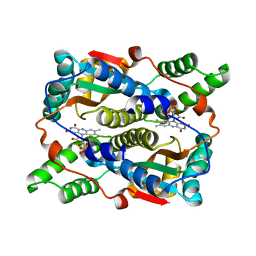 | | The structure of E.coli nitroreductase with bound acetate, crystal form 1 | | Descriptor: | ACETATE ION, FLAVIN MONONUCLEOTIDE, Oxygen-insensitive NAD(P)H nitroreductase | | Authors: | Race, P.R, Lovering, A.L, Green, R.M, Ossor, A, White, S.A, Searle, P.F, Wrighton, C.J, Hyde, E.I. | | Deposit date: | 2005-01-19 | | Release date: | 2005-02-08 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural and mechanistic studies of Escherichia coli nitroreductase with the antibiotic nitrofurazone. Reversed binding orientations in different redox states of the enzyme.
J.Biol.Chem., 280, 2005
|
|
6BR1
 
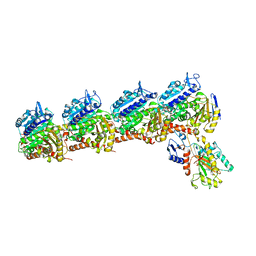 | | Tubulin-RB3_SLD-TTL in complex with heterocyclic pyrimidine compound 4a | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-chloro-4-(6-methoxy-3,4-dihydroquinolin-1(2H)-yl)pyrido[2,3-d]pyrimidine, CALCIUM ION, ... | | Authors: | Kumar, G, Wang, Y, Li, W, White, S.W. | | Deposit date: | 2017-11-29 | | Release date: | 2018-06-27 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.304 Å) | | Cite: | Heterocyclic-Fused Pyrimidines as Novel Tubulin Polymerization Inhibitors Targeting the Colchicine Binding Site: Structural Basis and Antitumor Efficacy.
J. Med. Chem., 61, 2018
|
|
5D4G
 
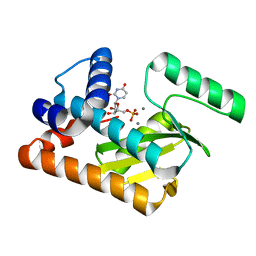 | | 2009 H1N1 PA endonuclease mutant F105S in complex with rUMP | | Descriptor: | MANGANESE (II) ION, Polymerase acidic protein, URIDINE-5'-MONOPHOSPHATE | | Authors: | Kumar, G, White, S.W. | | Deposit date: | 2015-08-07 | | Release date: | 2016-03-23 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Identification and characterization of influenza variants resistant to a viral endonuclease inhibitor.
Proc. Natl. Acad. Sci. U.S.A., 113, 2016
|
|
5DBS
 
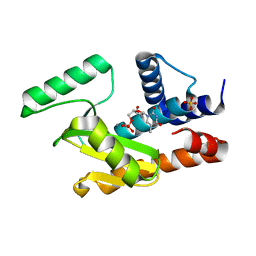 | | 2009 H1N1 PA endonuclease mutant E119D in complex with dTMP | | Descriptor: | MANGANESE (II) ION, Polymerase acidic protein, SULFATE ION, ... | | Authors: | Kumar, G, White, S.W. | | Deposit date: | 2015-08-21 | | Release date: | 2016-03-23 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Identification and characterization of influenza variants resistant to a viral endonuclease inhibitor.
Proc. Natl. Acad. Sci. U.S.A., 113, 2016
|
|
