6I4Q
 
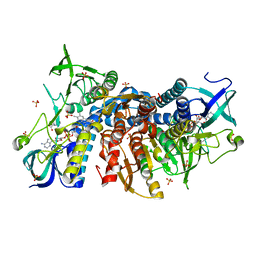 | | Crystal structure of the human dihydrolipoamide dehydrogenase at 1.75 Angstrom resolution | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Dihydrolipoyl dehydrogenase, mitochondrial, ... | | Authors: | Nagy, B, Szabo, E, Wilk, P, Torocsik, B, Weiss, M.S, Adam-Vizi, V, Ambrus, A. | | Deposit date: | 2018-11-10 | | Release date: | 2019-11-20 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Underlying molecular alterations in human dihydrolipoamide dehydrogenase deficiency revealed by structural analyses of disease-causing enzyme variants.
Hum.Mol.Genet., 28, 2019
|
|
6I4U
 
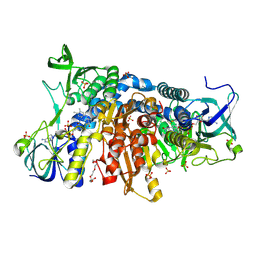 | | Crystal structure of the disease-causing G426E mutant of the human dihydrolipoamide dehydrogenase | | Descriptor: | Dihydrolipoyl dehydrogenase, mitochondrial, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Szabo, E, Wilk, P, Hubert, A, Torocsik, B, Weiss, M.S, Adam-Vizi, V, Ambrus, A. | | Deposit date: | 2018-11-10 | | Release date: | 2019-11-20 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Underlying molecular alterations in human dihydrolipoamide dehydrogenase deficiency revealed by structural analyses of disease-causing enzyme variants.
Hum.Mol.Genet., 28, 2019
|
|
6I4Z
 
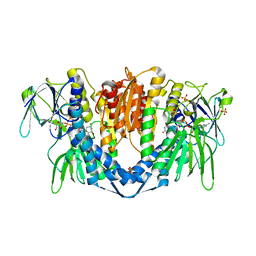 | | Crystal structure of the disease-causing P453L mutant of the human dihydrolipoamide dehydrogenase | | Descriptor: | Dihydrolipoyl dehydrogenase, mitochondrial, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Szabo, E, Wilk, P, Torocsik, B, Weiss, M.S, Adam-Vizi, V, Ambrus, A. | | Deposit date: | 2018-11-12 | | Release date: | 2019-11-20 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.342 Å) | | Cite: | Underlying molecular alterations in human dihydrolipoamide dehydrogenase deficiency revealed by structural analyses of disease-causing enzyme variants.
Hum.Mol.Genet., 28, 2019
|
|
6I4P
 
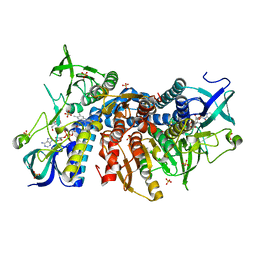 | | Crystal structure of the disease-causing G194C mutant of the human dihydrolipoamide dehydrogenase | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Dihydrolipoyl dehydrogenase, mitochondrial, ... | | Authors: | Szabo, E, Wilk, P, Torocsik, B, Weiss, M.S, Adam-Vizi, V, Ambrus, A. | | Deposit date: | 2018-11-10 | | Release date: | 2019-11-20 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Underlying molecular alterations in human dihydrolipoamide dehydrogenase deficiency revealed by structural analyses of disease-causing enzyme variants.
Hum.Mol.Genet., 28, 2019
|
|
6I4R
 
 | | Crystal structure of the disease-causing R460G mutant of the human dihydrolipoamide dehydrogenase at 1.44 Angstrom resolution | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Dihydrolipoyl dehydrogenase, mitochondrial, ... | | Authors: | Szabo, E, Wilk, P, Bui, D, Torocsik, B, Weiss, M.S, Adam-Vizi, V, Ambrus, A. | | Deposit date: | 2018-11-10 | | Release date: | 2019-11-20 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.439 Å) | | Cite: | Underlying molecular alterations in human dihydrolipoamide dehydrogenase deficiency revealed by structural analyses of disease-causing enzyme variants.
Hum.Mol.Genet., 28, 2019
|
|
6I4S
 
 | | Crystal structure of the disease-causing R447G mutant of the human dihydrolipoamide dehydrogenase | | Descriptor: | Dihydrolipoyl dehydrogenase, mitochondrial, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Szabo, E, Wilk, P, Hubert, A, Torocsik, B, Weiss, M.S, Adam-Vizi, V, Ambrus, A. | | Deposit date: | 2018-11-10 | | Release date: | 2019-11-20 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Underlying molecular alterations in human dihydrolipoamide dehydrogenase deficiency revealed by structural analyses of disease-causing enzyme variants.
Hum.Mol.Genet., 28, 2019
|
|
8DGD
 
 | | Crystal Structure of GAF domain-containing protein, from Klebsiella pneumoniae | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, GAF domain-containing protein, ZINC ION | | Authors: | Seattle Structural Genomics Center for Infectious Disease, Delker, S.L, Weiss, M.J, Lorimer, D.D, Edwards, T.E, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2022-06-23 | | Release date: | 2023-03-01 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure of GAF domain-containing protein, from Klebsiella pneumoniae
to be published
|
|
1Q4V
 
 | | CRYSTAL STRUCTURE OF ALLO-ILEA2-INSULIN, AN INACTIVE CHIRAL ANALOGUE: IMPLICATIONS FOR THE MECHANISM OF RECEPTOR | | Descriptor: | Insulin, PHENOL, ZINC ION | | Authors: | Wan, Z.L, Xu, B, Chu, Y.C, Katsoyannis, P.G, Weiss, M.A. | | Deposit date: | 2003-08-04 | | Release date: | 2003-08-19 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of allo-Ile(A2)-insulin, an inactive chiral analogue: implications for the
mechanism of receptor binding.
Biochemistry, 42, 2003
|
|
7NCX
 
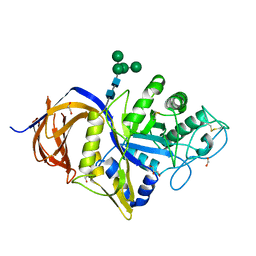 | | Crystal structure of GH30 (double mutant EE) from Thermothelomyces thermophila. | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Dimarogona, M, Nikolaivits, E, Topakas, E, Weiss, M, Feiler, C.G. | | Deposit date: | 2021-01-29 | | Release date: | 2021-09-08 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | Unique features of the bifunctional GH30 from Thermothelomyces thermophila revealed by structural and mutational studies
Carbohydrate Polymers, 273, 2021
|
|
5ZNF
 
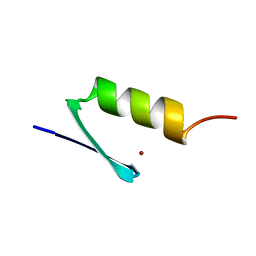 | |
7ZNF
 
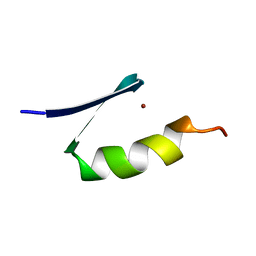 | |
4WUO
 
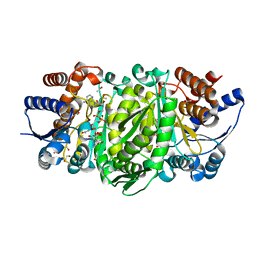 | | Structure of the E270A Mutant Isopropylmalate dehydrogenase from Thermus thermophilus in complex with IPM, Mn and NADH | | Descriptor: | 3-ISOPROPYLMALIC ACID, 3-isopropylmalate dehydrogenase, ETHANOL, ... | | Authors: | Pallo, A, Graczer, E, Olah, J, Szimler, T, Konarev, P.V, Svergun, D.I, Merli, A, Zavodszky, P, Vas, M, Weiss, M.S. | | Deposit date: | 2014-11-03 | | Release date: | 2014-11-12 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Glutamate 270 plays an essential role in K(+)-activation and domain closure of Thermus thermophilus isopropylmalate dehydrogenase.
Febs Lett., 589, 2015
|
|
6YNQ
 
 | | Structure of SARS-CoV-2 Main Protease bound to 2-Methyl-1-tetralone. | | Descriptor: | (2~{S})-2-methyl-3,4-dihydro-2~{H}-naphthalen-1-one, 3C-like proteinase, CHLORIDE ION, ... | | Authors: | Guenther, S, Reinke, P, Oberthuer, D, Yefanov, O, Gelisio, L, Ginn, H, Lieske, J, Domaracky, M, Brehm, W, Rahmani Mashour, A, White, T.A, Knoska, J, Pena Esperanza, G, Koua, F, Tolstikova, A, Groessler, M, Fischer, P, Hennicke, V, Fleckenstein, H, Trost, F, Galchenkova, M, Gevorkov, Y, Li, C, Awel, S, Paulraj, L.X, Ullah, N, Falke, S, Alves Franca, B, Schwinzer, M, Brognaro, H, Werner, N, Perbandt, M, Tidow, H, Seychell, B, Beck, T, Meier, S, Doyle, J.J, Giseler, H, Melo, D, Dunkel, I, Lane, T.J, Peck, A, Saouane, S, Hakanpaeae, J, Meyer, J, Noei, H, Gribbon, P, Ellinger, B, Kuzikov, M, Wolf, M, Zhang, L, Ehrt, C, Pletzer-Zelgert, J, Wollenhaupt, J, Feiler, C, Weiss, M, Schulz, E.C, Mehrabi, P, Norton-Baker, B, Schmidt, C, Lorenzen, K, Schubert, R, Han, H, Chari, A, Fernandez Garcia, Y, Turk, D, Hilgenfeld, R, Rarey, M, Zaliani, A, Chapman, H.N, Pearson, A, Betzel, C, Meents, A. | | Deposit date: | 2020-04-14 | | Release date: | 2020-04-29 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | X-ray screening identifies active site and allosteric inhibitors of SARS-CoV-2 main protease.
Science, 372, 2021
|
|
7O4F
 
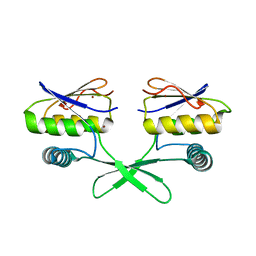 | | The DYW domain of A. thaliana OTP86 in its active state | | Descriptor: | Pentatricopeptide repeat-containing protein At3g63370, chloroplastic, ZINC ION | | Authors: | Weber, G, Palm, G.J, Takenaka, M, Barthel, T, Feiler, C, Weiss, M.S. | | Deposit date: | 2021-04-06 | | Release date: | 2021-06-30 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | DYW domain structures imply an unusual regulation principle in plant organellar RNA editing catalysis.
Nat Catal, 4, 2021
|
|
7O4E
 
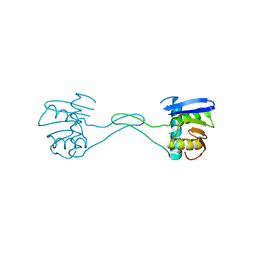 | | The DYW domain of A. thaliana OTP86 in its inactive state | | Descriptor: | Pentatricopeptide repeat-containing protein At3g63370, chloroplastic, ZINC ION | | Authors: | Weber, G, Palm, G.J, Takenaka, M, Barthel, T, Feiler, C, Weiss, M.S. | | Deposit date: | 2021-04-06 | | Release date: | 2021-06-30 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | DYW domain structures imply an unusual regulation principle in plant organellar RNA editing catalysis.
Nat Catal, 4, 2021
|
|
6YVF
 
 | | Structure of SARS-CoV-2 Main Protease bound to AZD6482. | | Descriptor: | 2-[[(1R)-1-(7-methyl-2-morpholin-4-yl-4-oxidanylidene-pyrido[1,2-a]pyrimidin-9-yl)ethyl]amino]benzoic acid, 3C-like proteinase, CALCIUM ION, ... | | Authors: | Guenther, S, Reinke, P, Oberthuer, D, Yefanov, O, Gelisio, L, Ginn, H, Lieske, J, Domaracky, M, Brehm, W, Rahmani Mashour, A, White, T.A, Knoska, J, Pena Esperanza, G, Koua, F, Tolstikova, A, Groessler, M, Fischer, P, Hennicke, V, Fleckenstein, H, Trost, F, Galchenkova, M, Gevorkov, Y, Li, C, Awel, S, Paulraj, L.X, Ullah, N, Falke, S, Alves Franca, B, Schwinzer, M, Brognaro, H, Werner, N, Perbandt, M, Tidow, H, Seychell, B, Beck, T, Meier, S, Doyle, J.J, Giseler, H, Melo, D, Dunkel, I, Lane, T.J, Peck, A, Saouane, S, Hakanpaeae, J, Meyer, J, Noei, H, Gribbon, P, Ellinger, B, Kuzikov, M, Wolf, M, Zhang, L, Ehrt, C, Pletzer-Zelgert, J, Wollenhaupt, J, Feiler, C, Weiss, M, Schulz, E.C, Mehrabi, P, Norton-Baker, B, Schmidt, C, Lorenzen, K, Schubert, R, Han, H, Chari, A, Fernandez Garcia, Y, Turk, D, Hilgenfeld, R, Rarey, M, Zaliani, A, Chapman, H.N, Pearson, A, Betzel, C, Meents, A. | | Deposit date: | 2020-04-28 | | Release date: | 2020-05-20 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | X-ray screening identifies active site and allosteric inhibitors of SARS-CoV-2 main protease.
Science, 372, 2021
|
|
3KQ6
 
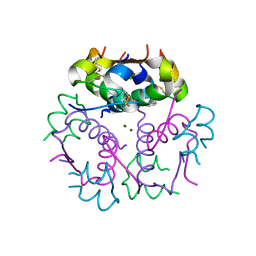 | | Enhancing the Therapeutic Properties of a Protein by a Designed Zinc-Binding Site, Structural principles of a novel long-acting insulin analog | | Descriptor: | CHLORIDE ION, Insulin A chain, Insulin B chain, ... | | Authors: | Wan, Z.L, Hu, S.Q, Whittaker, L, Phillips, N.B, Whittake, J, Ismail-Beigi, F, Weiss, M.A. | | Deposit date: | 2009-11-17 | | Release date: | 2010-02-23 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Supramolecular protein engineering: design of zinc-stapled insulin hexamers as a long acting depot.
J.Biol.Chem., 285, 2010
|
|
7ABU
 
 | | Structure of SARS-CoV-2 Main Protease bound to RS102895 | | Descriptor: | 1'-[2-[4-(trifluoromethyl)phenyl]ethyl]spiro[1~{H}-3,1-benzoxazine-4,4'-piperidine]-2-one, 3C-like proteinase, DIMETHYL SULFOXIDE, ... | | Authors: | Guenther, S, Reinke, P.Y.A, Oberthuer, D, Yefanov, O, Gelisio, L, Ginn, H, Lieske, J, Domaracky, M, Brehm, W, Rahmani Mashour, A, White, T.A, Knoska, J, Pena Esperanza, G, Koua, F, Tolstikova, A, Groessler, M, Fischer, P, Hennicke, V, Fleckenstein, H, Trost, F, Galchenkova, M, Gevorkov, Y, Li, C, Awel, S, Paulraj, L.X, Ullah, N, Falke, S, Alves Franca, B, Schwinzer, M, Brognaro, H, Werner, N, Perbandt, M, Tidow, H, Seychell, B, Beck, T, Meier, S, Doyle, J.J, Giseler, H, Melo, D, Lane, T.J, Dunkel, I, Peck, A, Saouane, S, Hakanpaeae, J, Meyer, J, Noei, H, Gribbon, P, Ellinger, B, Kuzikov, M, Wolf, M, Zhang, L, Ehrt, C, Pletzer-Zelgert, J, Wollenhaupt, J, Feiler, C, Weiss, M, Schulz, E.C, Mehrabi, P, Norton-Baker, B, Schmidt, C, Lorenzen, K, Schubert, R, Han, H, Chari, A, Fernandez Garcia, Y, Turk, D, Hilgenfeld, R, Rarey, M, Zaliani, A, Chapman, H.N, Pearson, A, Betzel, C, Meents, A. | | Deposit date: | 2020-09-08 | | Release date: | 2020-12-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | X-ray screening identifies active site and allosteric inhibitors of SARS-CoV-2 main protease.
Science, 372, 2021
|
|
7O0E
 
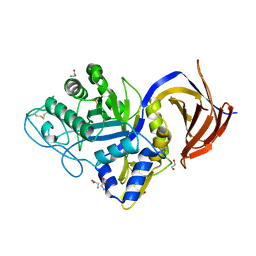 | | Crystal structure of GH30 (mutant E188A) complexed with aldotriuronic acid from Thermothelomyces thermophila. | | Descriptor: | 1,2-ETHANEDIOL, 4-O-methyl-alpha-D-glucopyranuronic acid-(1-2)-beta-D-xylopyranose-(1-4)-beta-D-xylopyranose, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Dimarogona, M, Kosinas, C, Feiler, C, Weiss, M.S, Topakas, E, Nikolaivits, E. | | Deposit date: | 2021-03-26 | | Release date: | 2021-09-08 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Unique features of the bifunctional GH30 from Thermothelomyces thermophila revealed by structural and mutational studies
Carbohydrate Polymers, 273, 2021
|
|
1B4T
 
 | | H48C YEAST CU(II)/ZN SUPEROXIDE DISMUTASE ROOM TEMPERATURE (298K) STRUCTURE | | Descriptor: | CHLORIDE ION, COPPER (II) ION, PROTEIN (CU/ZN SUPEROXIDE DISMUTASE), ... | | Authors: | Hart, P.J, Balbirnie, M.M, Ogihara, N.L, Nersissian, A.M, Weiss, M.S, Valentine, J.S, Eisenberg, D. | | Deposit date: | 1998-12-23 | | Release date: | 1999-12-23 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A structure-based mechanism for copper-zinc superoxide dismutase.
Biochemistry, 38, 1999
|
|
1B4L
 
 | | 15 ATMOSPHERE OXYGEN YEAST CU/ZN SUPEROXIDE DISMUTASE ROOM TEMPERATURE (298K) STRUCTURE | | Descriptor: | COPPER (II) ION, PROTEIN (CU/ZN SUPEROXIDE DISMUTASE), ZINC ION | | Authors: | Hart, P.J, Balbirnie, M.M, Ogihara, N.L, Nersissian, A.M, Weiss, M.S, Valentine, J.S, Eisenberg, D. | | Deposit date: | 1998-12-22 | | Release date: | 1999-12-23 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A structure-based mechanism for copper-zinc superoxide dismutase.
Biochemistry, 38, 1999
|
|
3P2X
 
 | | Insulin fibrillation is the Janus face of induced fit. A chiaral clamp stabilizes the native state at the expense of activity | | Descriptor: | CHLORIDE ION, Insulin, PHENOL, ... | | Authors: | Hua, Q.X, Wan, Z.L, Huang, K, Hu, S.Q, Phillip, N.F, Jia, W.H, Whittingham, J, Dodson, G.G, Katsoyannis, P.G, Weiss, M.A. | | Deposit date: | 2010-10-04 | | Release date: | 2011-11-23 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Insulin fibrillation is the Janus face of induced fit. A chiral clamp stabilizes the native state at the expense of activity
To be Published
|
|
3P33
 
 | | Insulin fibrillation is the Janus face of induced fit. A chiral clamp stabilizes the native state at the expense of activity | | Descriptor: | CHLORIDE ION, Insulin, PHENOL, ... | | Authors: | Hua, Q.X, Wan, Z.L, Huang, K, Hu, S.Q, Phillip, N.F, Jia, W.H, Whittingham, J, Dodson, G.G, Katsoyannis, P.G, Weiss, M.A. | | Deposit date: | 2010-10-04 | | Release date: | 2011-11-23 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Insulin fibrillation is the Janus face of induced fit. A chiral clamp stabilizes the native state at the expense of activity
To be Published
|
|
3V1G
 
 | | Forestalling insulin fibrillation by insertion of a chiral clamp mechanism-based application of protein engineering to global health | | Descriptor: | CHLORIDE ION, Insulin, PHENOL, ... | | Authors: | Wan, Z.L, Hua, Q.X, Wickramasinghe, N.P, Huang, K, Petkova, A.T, Hu, S.Q, Phillips, N.B, Yeh, I.J, Whittake, J, Ismail-Beigi, F, Katsoyyannis, P.G, Tycko, R, Weiss, M.A. | | Deposit date: | 2011-12-09 | | Release date: | 2012-12-12 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Forestalling insulin fibrillation by insertion of a
chiral clamp mechanism-based application of protein engineering to global health
To be Published
|
|
3V19
 
 | | Forestalling insulin fibrillation by insertion of a chiral clamp mechanism-based application of protein engineering to global health | | Descriptor: | CHLORIDE ION, Insulin, PHENOL, ... | | Authors: | Wan, Z.L, Hua, Q.X, Wickramasinghe, N.P, Huang, K, Petkova, A.T, Hu, S.Q, Phillips, N.B, Yeh, I.J, Whittake, J, Ismail-Beigi, F, Katsoyyannis, P.G, Tycko, R, Weiss, M.A. | | Deposit date: | 2011-12-09 | | Release date: | 2012-12-12 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Forestalling insulin fibrillation by insertion of a
chiral clamp mechanism-based application of protein engineering to global health
To be Published
|
|
