3SMR
 
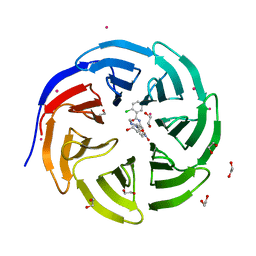 | | Crystal structure of human WD repeat domain 5 with compound | | Descriptor: | 1,2-ETHANEDIOL, 2-chloro-N-[2-(4-methylpiperazin-1-yl)-5-nitrophenyl]benzamide, UNKNOWN ATOM OR ION, ... | | Authors: | Dong, A, Dombrovski, L, Wasney, G.A, Tempel, W, Senisterra, G, Bolshan, Y, Smil, D, Nguyen, K.T, Hajian, T, Poda, G, Al-Awar, R, Bountra, C, Weigelt, J, Edwards, A.M, Brown, P.J, Schapira, M, Arrowsmith, C.H, Vedadi, M, Wu, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2011-06-28 | | Release date: | 2011-08-31 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Small-molecule inhibition of MLL activity by disruption of its interaction with WDR5.
Biochem. J., 449, 2013
|
|
4ABL
 
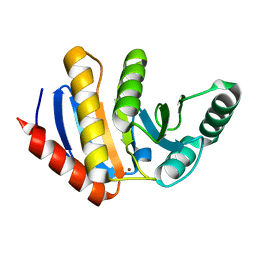 | | HUMAN PARP14 (ARTD8, BAL2) - MACRO DOMAIN 3 | | Descriptor: | BROMIDE ION, POLY [ADP-RIBOSE] POLYMERASE 14 | | Authors: | Karlberg, T, Moche, M, Thorsell, A.G, Arrowsmith, C.H, Bountra, C, Edwards, A.M, Ekblad, T, Weigelt, J, Schuler, H. | | Deposit date: | 2011-12-08 | | Release date: | 2012-12-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Recognition of Mono-Adp-Ribosylated Artd10 Substrates by Artd8 Macrodomains
Structure, 21, 2013
|
|
4A9C
 
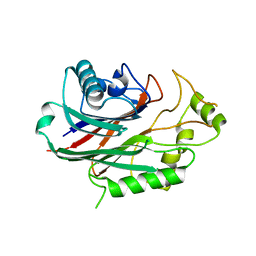 | | Crystal structure of human SHIP2 in complex with biphenyl 2,3',4,5',6- pentakisphosphate | | Descriptor: | BIPHENYL 2,3',4,5',6-PENTAKISPHOSPHATE, PHOSPHATIDYLINOSITOL-3,4,5-TRISPHOSPHATE 5-PHOSPHATASE 2 | | Authors: | Tresaugues, L, Arrowsmith, C.H, Berglund, H, Bountra, C, Edwards, A.M, Ekblad, T, Graslund, S, Karlberg, T, Mills, S.J, Moche, M, Nyman, T, Persson, C, Potter, B.V.L, Schuler, H, Thorsell, A.G, Weigelt, J, Nordlund, P. | | Deposit date: | 2011-11-25 | | Release date: | 2012-05-30 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A Synthetic Polyphosphoinositide Headgroup Surrogate in Complex with Ship2 Provides a Rationale for Drug Discovery.
Acs Chem.Biol., 7, 2012
|
|
3LJU
 
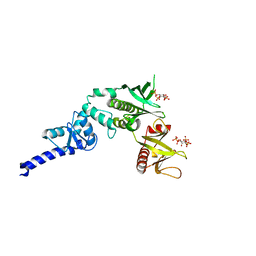 | | Crystal structure of full length centaurin alpha-1 bound with the head group of PIP3 | | Descriptor: | (2R)-3-{[(R)-{[(1S,2S,3R,4S,5S,6S)-2,6-dihydroxy-3,4,5-tris(phosphonooxy)cyclohexyl]oxy}(hydroxy)phosphoryl]oxy}propane -1,2-diyl dioctanoate, Arf-GAP with dual PH domain-containing protein 1, ZINC ION | | Authors: | Shen, L, Tong, Y, Tempel, W, MacKenzie, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Weigelt, J, Bochkarev, A, Park, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-01-26 | | Release date: | 2010-11-24 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.702 Å) | | Cite: | Phosphorylation-independent dual-site binding of the FHA domain of KIF13 mediates phosphoinositide transport via centaurin alpha1.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
4ABK
 
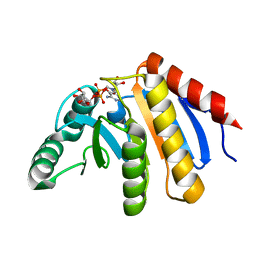 | | HUMAN PARP14 (ARTD8, BAL2) - MACRO DOMAIN 3 IN COMPLEX WITH ADENOSINE- 5-DIPHOSPHORIBOSE | | Descriptor: | POLY [ADP-RIBOSE] POLYMERASE 14, [(2R,3S,4R,5R)-5-(6-AMINOPURIN-9-YL)-3,4-DIHYDROXY-OXOLAN-2-YL]METHYL [HYDROXY-[[(2R,3S,4R,5S)-3,4,5-TRIHYDROXYOXOLAN-2-YL]METHOXY]PHOSPHORYL] HYDROGEN PHOSPHATE | | Authors: | Karlberg, T, Moche, M, Thorsell, A.G, Arrowsmith, C.H, Bountra, C, Edwards, A.M, Ekblad, T, Weigelt, J, Schuler, H. | | Deposit date: | 2011-12-08 | | Release date: | 2012-12-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Recognition of Mono-Adp-Ribosylated Artd10 Substrates by Artd8 Macrodomains
Structure, 21, 2013
|
|
3LD6
 
 | | Crystal structure of human lanosterol 14alpha-demethylase (CYP51) in complex with ketoconazole | | Descriptor: | 1-acetyl-4-(4-{[(2R,4S)-2-(2,4-dichlorophenyl)-2-(1H-imidazol-1-ylmethyl)-1,3-dioxolan-4-yl]methoxy}phenyl)piperazine, Cycloheptakis-(1-4)-(alpha-D-glucopyranose), Lanosterol 14-alpha demethylase, ... | | Authors: | Strushkevich, N, Tempel, W, MacKenzie, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Weigelt, J, Park, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-01-12 | | Release date: | 2010-02-02 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural basis of human CYP51 inhibition by antifungal azoles.
J.Mol.Biol., 397, 2010
|
|
3LLM
 
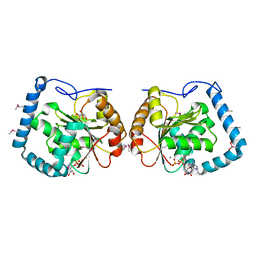 | | Crystal Structure Analysis of a RNA Helicase | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP-dependent RNA helicase A, CACODYLATE ION, ... | | Authors: | Schutz, P, Karlberg, T, Collins, R, Arrowsmith, C.H, Berglund, H, Bountra, C, Flodin, S, Flores, A, Graslund, S, Hammarstrom, M, Johansson, A, Johansson, I, Kallas, A, Kraulis, P, Kotenyova, T, Kotzsch, A, Markova, N, Moche, M, Nielsen, T.K, Nordlund, P, Nyman, T, Persson, C, Roos, A.K, Siponen, M.I, Svensson, L, Thorsell, A.G, Tresaugues, L, Van Den Berg, S, Wahlberg, E, Weigelt, J, Welin, M, Wisniewska, M, Schuler, H.M, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-01-29 | | Release date: | 2010-05-12 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of human RNA helicase A (DHX9): structural basis for unselective nucleotide base binding in a DEAD-box variant protein.
J.Mol.Biol., 400, 2010
|
|
3LQ3
 
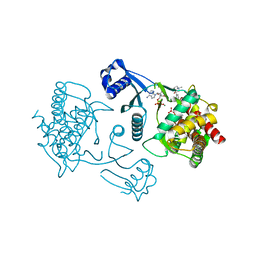 | | Crystal structure of human choline kinase beta in complex with phosphorylated hemicholinium-3 and adenosine nucleotide | | Descriptor: | (2S)-2-[4'-({dimethyl[2-(phosphonooxy)ethyl]ammonio}acetyl)biphenyl-4-yl]-2-hydroxy-4,4-dimethylmorpholin-4-ium, ADENOSINE MONOPHOSPHATE, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Hong, B.S, Tempel, W, Rabeh, W.M, MacKenzie, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Weigelt, J, Bochkarev, A, Park, H.W, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-02-08 | | Release date: | 2010-05-05 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Crystal structures of human choline kinase isoforms in complex with hemicholinium-3: single amino acid near the active site influences inhibitor sensitivity.
J.Biol.Chem., 285, 2010
|
|
3UVW
 
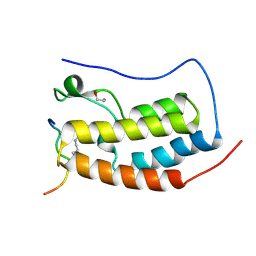 | | Crystal Structure of the first bromodomain of human BRD4 in complex with a diacetylated histone 4 peptide (H4K5acK8ac) | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain-containing protein 4, Histone H4 | | Authors: | Filippakopoulos, P, Felletar, I, Picaud, S, Keates, T, Muniz, J, Gileadi, O, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Weigelt, J, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2011-11-30 | | Release date: | 2012-01-18 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.37 Å) | | Cite: | Histone recognition and large-scale structural analysis of the human bromodomain family.
Cell(Cambridge,Mass.), 149, 2012
|
|
3UW9
 
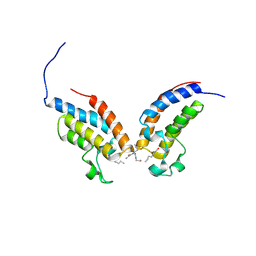 | | Crystal Structure of the first bromodomain of human BRD4 in complex with a diacetylated histone 4 peptide (H4K8acK12ac) | | Descriptor: | Bromodomain-containing protein 4, histone 4 peptide (H4K8acK12ac) | | Authors: | Filippakopoulos, P, Felletar, I, Picaud, S, Keates, T, Muniz, J, Gileadi, O, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Weigelt, J, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2011-12-01 | | Release date: | 2012-03-14 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Histone recognition and large-scale structural analysis of the human bromodomain family.
Cell(Cambridge,Mass.), 149, 2012
|
|
3UV5
 
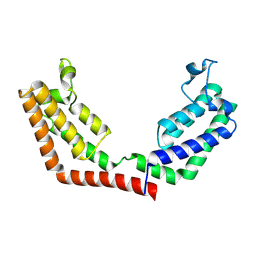 | | Crystal Structure of the tandem bromodomains of human Transcription initiation factor TFIID subunit 1 (TAF1) | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Transcription initiation factor TFIID subunit 1 | | Authors: | Filippakopoulos, P, Felletar, I, Picaud, S, Keates, T, Muniz, J, Gileadi, O, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Weigelt, J, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2011-11-29 | | Release date: | 2012-01-18 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Histone recognition and large-scale structural analysis of the human bromodomain family.
Cell(Cambridge,Mass.), 149, 2012
|
|
3UVY
 
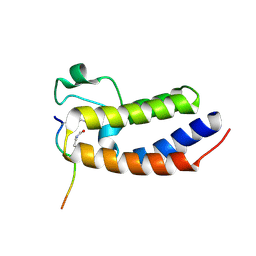 | | Crystal Structure of the first bromodomain of human BRD4 in complex with a diacetylated histone 4 peptide (H4K16acK20ac) | | Descriptor: | Bromodomain-containing protein 4, Histone H4 | | Authors: | Filippakopoulos, P, Felletar, I, Picaud, S, Keates, T, Muniz, J, Gileadi, O, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Weigelt, J, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2011-11-30 | | Release date: | 2012-01-18 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Histone recognition and large-scale structural analysis of the human bromodomain family.
Cell(Cambridge,Mass.), 149, 2012
|
|
3LYR
 
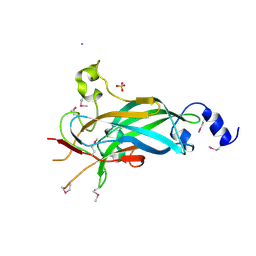 | | Human Early B-cell Factor 1 (EBF1) DNA-binding domain | | Descriptor: | AMMONIUM ION, SULFATE ION, Transcription factor COE1, ... | | Authors: | Siponen, M.I, Wisniewska, M, Arrowsmith, C.H, Bountra, C, Collins, R, Edwards, A.M, Flodin, S, Flores, A, Graslund, S, Hammarstrom, M, Johansson, I, Karlberg, T, Kraulis, P, Kotenyova, T, Markova, N, Moche, M, Nordlund, P, Nyman, T, Persson, C, Schuler, H, Schutz, P, Svensson, L, Thorsell, A.G, Tresaugues, L, Van Den Berg, S, Wahlberg, E, Weigelt, J, Welin, M, Berglund, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-02-28 | | Release date: | 2010-03-16 | | Last modified: | 2014-10-08 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Structural Determination of Functional Domains in Early B-cell Factor (EBF) Family of Transcription Factors Reveals Similarities to Rel DNA-binding Proteins and a Novel Dimerization Motif.
J.Biol.Chem., 285, 2010
|
|
3LY5
 
 | | DDX18 dead-domain | | Descriptor: | ATP-dependent RNA helicase DDX18, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Schutz, P, Karlberg, T, Arrowsmith, C.H, Berglund, H, Bountra, C, Collins, R, Edwards, A.M, Flodin, S, Flores, A, Graslund, S, Hammarstrom, M, Johansson, A, Johansson, I, Kallas, A, Kraulis, P, Kotenyova, T, Kotzsch, A, Markova, N, Moche, M, Nielsen, T.K, Nordlund, P, Nyman, T, Persson, C, Roos, A.K, Siponen, M.I, Svensson, L, Thorsell, A.G, Tresaugues, L, Van Den Berg, S, Wahlberg, E, Weigelt, J, Welin, M, Wisniewska, M, Schuler, H.M, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-02-26 | | Release date: | 2010-05-12 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Comparative Structural Analysis of Human DEAD-Box RNA Helicases.
Plos One, 5, 2010
|
|
2BZN
 
 | | Crystal structure of human guanosine monophosphate reductase 2 GMPR2 in complex with IMP | | Descriptor: | GMP REDUCTASE 2, INOSINIC ACID | | Authors: | Stenmark, P, Kursula, P, Arrowsmith, C, Berglund, H, Edwards, A, Ehn, M, Graslund, S, Hammarstrom, M, Hallberg, B.M, Kotenyova, T, Nilsson-Ehle, P, Nordlund, P, Ogg, D, Persson, C, Sagemark, J, Schuler, H, Sundstrom, M, Thorsell, A, Weigelt, J. | | Deposit date: | 2005-08-19 | | Release date: | 2005-09-02 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Cofactor Mobility Determines Reaction Outcome in the Impdh and Gmpr (Beta-Alpha)(8) Barrel Enzymes.
Nat.Chem.Biol., 7, 2011
|
|
4AD9
 
 | | Crystal structure of human LACTB2. | | Descriptor: | 1,2-ETHANEDIOL, BETA-LACTAMASE-LIKE PROTEIN 2, ZINC ION | | Authors: | Allerston, C.K, Krojer, T, Shrestha, B, Burgess Brown, N, Chalk, R, Elkins, J.M, Filippakopoulos, P, Pike, A.C.W, Muniz, J.R.C, Vollmar, M, Arrowsmith, C.H, Weigelt, J, Edwards, A, Bountra, C, von Delft, F, Gileadi, O. | | Deposit date: | 2011-12-22 | | Release date: | 2012-02-01 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Identification of Lactb2, a Metallo-Beta-Lactamase Protein, as a Human Mitochondrial Endoribonuclease
Nucleic Acids Res., 44, 2016
|
|
3UVD
 
 | | Crystal Structure of the bromodomain of human Transcription activator BRG1 (SMARCA4) in complex with N-Methyl-2-pyrrolidone | | Descriptor: | 1-methylpyrrolidin-2-one, Transcription activator BRG1 | | Authors: | Filippakopoulos, P, Felletar, I, Picaud, S, Keates, T, Muniz, J, Gileadi, O, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Weigelt, J, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2011-11-29 | | Release date: | 2012-01-18 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Histone recognition and large-scale structural analysis of the human bromodomain family.
Cell(Cambridge,Mass.), 149, 2012
|
|
3K2O
 
 | | Structure of an oxygenase | | Descriptor: | ACETATE ION, Bifunctional arginine demethylase and lysyl-hydroxylase JMJD6, CHLORIDE ION, ... | | Authors: | Krojer, T, McDonough, M.A, Clifton, I.J, Mantri, M, Ng, S.S, Pike, A.C.W, Butler, D.S, Webby, C.J, Kochan, G, Bhatia, C, Bray, J.E, Chaikuad, A, Gileadi, O, von Delft, F, Weigelt, J, Arrowsmith, C.H, Bountra, C, Edwards, A.M, Schofield, C.J, Kavanagh, K.L, Oppermann, U, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-09-30 | | Release date: | 2009-11-03 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal Structure of the 2-Oxoglutarate- and Fe(II)-Dependent Lysyl Hydroxylase JMJD6.
J.Mol.Biol., 401, 2010
|
|
2BW0
 
 | | Crystal Structure of the hydrolase domain of Human 10-Formyltetrahydrofolate 2 dehydrogenase | | Descriptor: | 10-FORMYLTETRAHYDROFOLATE DEHYDROGENASE, SULFATE ION | | Authors: | Ogg, D.J, Stenmark, P, Arrowsmith, C, Edwards, A, Ehn, M, Graslund, S, Hammarstrom, M, Hallberg, M, Kotenyova, T, Nilsson-Ehle, P, Nordlund, P, Persson, C, Sagemark, J, Schuler, H, Sundstrom, M, Thorsell, A, Dobritzsch, D, Weigelt, J. | | Deposit date: | 2005-07-07 | | Release date: | 2005-07-08 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structures of the Hydrolase Domain of Human 10-Formyltetrahydrofolate Dehydrogenase and its Complex with a Substrate Analogue.
Acta Crystallogr.,Sect.D, 62, 2006
|
|
4ATM
 
 | | Crystal structure of the BAR domain of human Amphiphysin, isoform 1 at 1.8 Angstrom resolution featuring increased order at the N- terminus. | | Descriptor: | 1,2-ETHANEDIOL, AMPHIPHYSIN, GLYCEROL | | Authors: | Allerston, C.K, Krojer, T, Arrowsmith, C.H, Weigelt, J, Edwards, A, Bountra, C, von Delft, F, Gileadi, O. | | Deposit date: | 2012-05-08 | | Release date: | 2012-05-30 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.783 Å) | | Cite: | Crystal Structure of the Bar Domain of Human Amphiphysin, Isoform 1
To be Published
|
|
3JXU
 
 | | Crystal structure of the human 70kDa heat shock protein 1A (Hsp70-1) ATPase domain in complex with ADP and inorganic phosphate | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Heat shock 70 kDa protein 1, MAGNESIUM ION, ... | | Authors: | Wisniewska, M.M, Karlberg, T, Arrowsmith, C.H, Berglund, H, Bountra, C, Collins, R, Edwards, A.M, Flodin, S, Flores, A, Graslund, S, Hammarstrom, M, Johansson, A, Johansson, I, Kallas, A, Kraulis, P, Kotenyova, T, Kotzsch, A, Markova, N, Moche, M, Nielsen, T.K, Nordlund, P, Nyman, T, Persson, C, Roos, A.K, Siponen, M.I, Schutz, P, Svensson, L, Thorsell, A.G, Tresaugues, L, Van Den Berg, S, Wahlberg, E, Weigelt, J, Welin, M, Schuler, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-09-21 | | Release date: | 2009-10-20 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Crystal structures of the ATPase domains of four human Hsp70 isoforms: HSPA1L/Hsp70-hom, HSPA2/Hsp70-2, HSPA6/Hsp70B', and HSPA5/BiP/GRP78
Plos One, 5, 2010
|
|
3KJD
 
 | | Human poly(ADP-ribose) polymerase 2, catalytic fragment in complex with an inhibitor ABT-888 | | Descriptor: | (2R)-2-(7-carbamoyl-1H-benzimidazol-2-yl)-2-methylpyrrolidinium, GLYCEROL, Poly [ADP-ribose] polymerase 2 | | Authors: | Karlberg, T, Schutz, P, Arrowsmith, C.H, Berglund, H, Bountra, C, Collins, R, Edwards, A.M, Flodin, S, Flores, A, Graslund, S, Hammarstrom, M, Johansson, A, Johansson, I, Kallas, A, Kotenyova, T, Kotzsch, A, Kraulis, P, Nielsen, T.K, Moche, M, Nordlund, P, Nyman, T, Persson, C, Roos, A.K, Siponen, M.I, Thorsell, A.G, Tresaugues, L, Van Den Berg, S, Weigelt, J, Welin, M, Wisniewska, M, Schuler, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-11-03 | | Release date: | 2009-11-17 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structure of the catalytic domain of human PARP2 in complex with PARP inhibitor ABT-888.
Biochemistry, 49, 2010
|
|
2C6Q
 
 | | Crystal structure of human guanosine monophosphate reductase 2 GMPR2 in complex with IMP and NADPH | | Descriptor: | GMP REDUCTASE 2, INOSINIC ACID, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Kursula, P, Stenmark, P, Arrowsmith, C, Berglund, H, Edwards, A, Ehn, M, Graslund, S, Hammarstrom, M, Hallberg, B.M, Kotenyova, T, Nilsson-Ehle, P, Ogg, D, Persson, C, Sagemark, J, Schuler, H, Sundstrom, M, Thorsell, A, Weigelt, J, Nordlund, P. | | Deposit date: | 2005-11-11 | | Release date: | 2005-11-30 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Cofactor Mobility Determines Reaction Outcome in the Impdh and Gmpr (Beta-Alpha)(8) Barrel Enzymes.
Nat.Chem.Biol., 7, 2011
|
|
3KHU
 
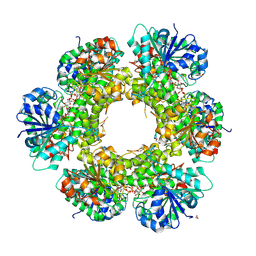 | | Crystal structure of human UDP-glucose dehydrogenase Glu161Gln, in complex with thiohemiacetal intermediate | | Descriptor: | 1,2-ETHANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, UDP-glucose 6-dehydrogenase, ... | | Authors: | Chaikuad, A, Egger, S, Yue, W.W, Guo, K, Sethi, R, Filippakopoulos, P, Muniz, J.R.C, von Delft, F, Bountra, C, Arrowsmith, C.H, Weigelt, J, Edwards, A.M, Kavanagh, K.L, Nidetzky, B, Oppermann, U, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-10-30 | | Release date: | 2009-11-17 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural and Kinetic Evidence That Catalytic Reaction of Human UDP-glucose 6-Dehydrogenase Involves Covalent Thiohemiacetal and Thioester Enzyme Intermediates.
J.Biol.Chem., 287, 2012
|
|
2CL3
 
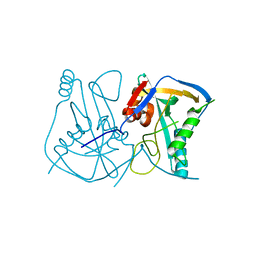 | | Crystal structure of human Cleavage and Polyadenylation Specificity Factor 5 (CPSF5) | | Descriptor: | CLEAVAGE AND POLYADENYLATION SPECIFICITY FACTOR 5 | | Authors: | Stenmark, P, Hogbom, M, Arrowsmith, C, Berglund, H, Collins, R, Edwards, A, Ehn, M, Flodin, S, Flores, A, Graslund, S, Hammarstrom, M, Hallberg, B.M, Holmberg Schiavone, L, Kotenyova, T, Magnusdottir, A, Nilsson-Ehle, P, Nyman, T, Ogg, D, Persson, C, Sagemark, J, Sundstrom, M, Thorsell, A.G, Van Den Berg, S, Wallden, K, Weigelt, J, Nordlund, P. | | Deposit date: | 2006-04-25 | | Release date: | 2006-05-04 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The Crystal Structure of Human Cleavage and Polyadenylation Specific Factor-5 Reveals a Dimeric Nudix Protein with a Conserved Catalytic Site.
Proteins, 73, 2008
|
|
