3EWR
 
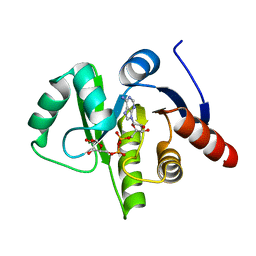 | | complex of substrate ADP-ribose with HCoV-229E Nsp3 ADRP domain | | Descriptor: | ADENOSINE-5-DIPHOSPHORIBOSE, Non-structural protein 3 | | Authors: | Xu, Y, Cong, L, Chen, C, Wei, L, Zhao, Q, Xu, X, Ma, Y, Bartlam, M, Rao, Z. | | Deposit date: | 2008-10-16 | | Release date: | 2009-01-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Crystal structures of two coronavirus ADP-ribose-1''-monophosphatases and their complexes with ADP-Ribose: a systematic structural analysis of the viral ADRP domain.
J.Virol., 83, 2009
|
|
3EWO
 
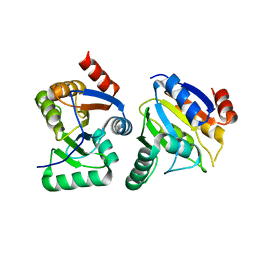 | | IBV Nsp3 ADRP domain | | Descriptor: | Non-structural protein 3 | | Authors: | Xu, Y, Cong, L, Chen, C, Wei, L, Zhao, Q, Xu, X, Ma, Y, Bartlam, M, Rao, Z. | | Deposit date: | 2008-10-16 | | Release date: | 2009-01-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structures of two coronavirus ADP-ribose-1''-monophosphatases and their complexes with ADP-Ribose: a systematic structural analysis of the viral ADRP domain.
J.Virol., 83, 2009
|
|
3EWP
 
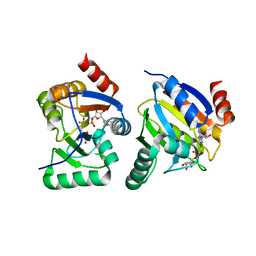 | | complex of substrate ADP-ribose with IBV Nsp3 ADRP domain | | Descriptor: | ADENOSINE-5-DIPHOSPHORIBOSE, Non-structural protein 3 | | Authors: | Xu, Y, Cong, L, Chen, C, Wei, L, Zhao, Q, Xu, X, Ma, Y, Bartlam, M, Rao, Z. | | Deposit date: | 2008-10-16 | | Release date: | 2009-01-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structures of two coronavirus ADP-ribose-1''-monophosphatases and their complexes with ADP-Ribose: a systematic structural analysis of the viral ADRP domain.
J.Virol., 83, 2009
|
|
4E1S
 
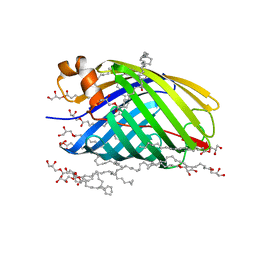 | | X-ray crystal structure of the transmembrane beta-domain from intimin from EHEC strain O157:H7 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (2S)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, CHLORIDE ION, ... | | Authors: | Fairman, J.W, Dautin, N, Wojtowicz, D, Wei, L, Noinaj, N, Barnard, T.J, Udho, E, Finkelstein, A, Przytycka, T.M, Cherezov, V, Buchanan, S.K. | | Deposit date: | 2012-03-07 | | Release date: | 2012-06-13 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.855 Å) | | Cite: | Crystal Structures of the Outer Membrane Domain of Intimin and Invasin from Enterohemorrhagic E. coli and Enteropathogenic Y. pseudotuberculosis.
Structure, 20, 2012
|
|
4E1T
 
 | | X-ray crystal structure of the transmembrane beta-domain from invasin from Yersinia pseudotuberculosis | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (2S)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Invasin | | Authors: | Fairman, J.W, Dautin, N, Wojtowicz, D, Wei, L, Noinaj, N, Barnard, T.J, Udho, E, Finkelstein, A, Przytycka, T.M, Cherezov, V, Buchanan, S.K. | | Deposit date: | 2012-03-07 | | Release date: | 2012-06-13 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.263 Å) | | Cite: | Crystal Structures of the Outer Membrane Domain of Intimin and Invasin from Enterohemorrhagic E. coli and Enteropathogenic Y. pseudotuberculosis.
Structure, 20, 2012
|
|
2IC1
 
 | | Crystal Structure of Human Cysteine Dioxygenase in Complex with Substrate Cysteine | | Descriptor: | CYSTEINE, Cysteine dioxygenase type 1, FE (II) ION | | Authors: | Ye, S, Wu, X, Wei, L, Tang, D, Sun, P, Rao, Z. | | Deposit date: | 2006-09-12 | | Release date: | 2006-12-05 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | An Insight into the Mechanism of Human Cysteine Dioxygenase: KEY ROLES OF THE THIOETHER-BONDED TYROSINE-CYSTEINE COFACTOR.
J.Biol.Chem., 282, 2007
|
|
7B38
 
 | | Torpedo californica acetylcholinesterase complexed with Mg+2 | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, Acetylcholinesterase, ... | | Authors: | Silman, I, Shnyrov, V.L, Ashani, Y, Roth, E, Nicolas, A, Sussman, J.L, Weiner, L. | | Deposit date: | 2020-11-29 | | Release date: | 2021-03-17 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Torpedo californica acetylcholinesterase is stabilized by binding of a divalent metal ion to a novel and versatile 4D motif.
Protein Sci., 30, 2021
|
|
7RK7
 
 | | The complex between TIL 1383i TCR and human Class I MHC HLA-A2 with the bound Tyrosinase(369-377)(N371D) nonameric peptide | | Descriptor: | Beta-2-microglobulin, HLA class I histocompatibility antigen, A alpha chain, ... | | Authors: | Singh, N.K, Davancaze, L.M, Arbuiso, A, Weiss, L.I, Keller, G.L.J, Baker, B.M. | | Deposit date: | 2021-07-22 | | Release date: | 2022-11-02 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.54 Å) | | Cite: | The complex between TIL 1383i TCR and human Class I MHC HLA-A2 with the bound Tyrosinase(369-377)(N371D) nonameric peptide
To Be Published
|
|
7B2W
 
 | | Torpedo californica acetylcholinesterase complexed with UO2 | | Descriptor: | Acetylcholinesterase, URANYL (VI) ION | | Authors: | Silman, I, Shnyrov, V.L, Ashani, Y, Roth, E, Nicolas, A, Sussman, J.L, Weiner, L. | | Deposit date: | 2020-11-28 | | Release date: | 2021-03-17 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Torpedo californica acetylcholinesterase is stabilized by binding of a divalent metal ion to a novel and versatile 4D motif.
Protein Sci., 30, 2021
|
|
6X5A
 
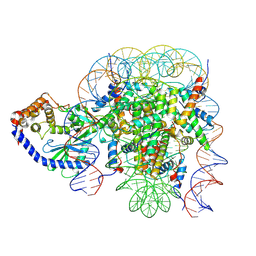 | | The mouse cGAS catalytic domain binding to human nucleosome that purified from HEK293T cells | | Descriptor: | Cyclic GMP-AMP synthase, DNA (natural), Histone H2A type 1, ... | | Authors: | Pengbiao, X, Pingwei, L, Baoyu, Z. | | Deposit date: | 2020-05-25 | | Release date: | 2020-09-16 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (4.36 Å) | | Cite: | The molecular basis of tight nuclear tethering and inactivation of cGAS.
Nature, 587, 2020
|
|
9IY0
 
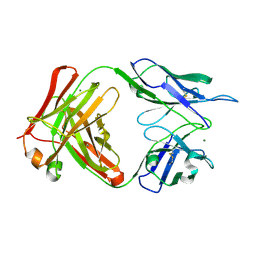 | | anti-HEV mAb 8H3 | | Descriptor: | Heavy chain of 8H3 Fab, Light chain of 8H3 Fab, MAGNESIUM ION | | Authors: | Minghua, Z, Lizhi, Z, Yang, H, Ying, G, Shaowei, L. | | Deposit date: | 2024-07-29 | | Release date: | 2024-09-04 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Structural basis for the synergetic neutralization of hepatitis E virus by antibody-antibody interaction
To Be Published
|
|
9IY2
 
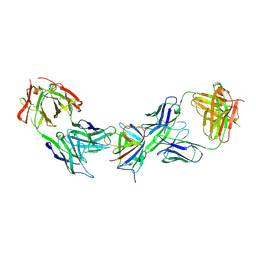 | | Immune complex of HEV-E2s, nAb 8C11 and nAb 8H3 | | Descriptor: | Heavy Chain of mAb 8C11, Heavy Chain of mAb 8H3, Light Chain of mAb 8C11, ... | | Authors: | Minghua, Z, Lizhi, Z, Ying, G, Shaowei, L. | | Deposit date: | 2024-07-29 | | Release date: | 2024-08-28 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.476 Å) | | Cite: | Structural basis for the synergetic neutralization of hepatitis E virus by antibody-antibody interaction
To Be Published
|
|
6X59
 
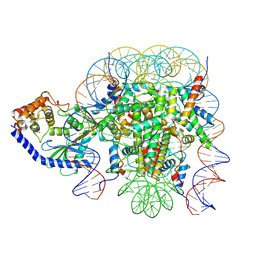 | | The mouse cGAS catalytic domain binding to human assembled nucleosome | | Descriptor: | Cyclic GMP-AMP synthase, DNA, Histone H2A type 1, ... | | Authors: | Pengbiao, X, Pingwei, L, Baoyu, Z. | | Deposit date: | 2020-05-25 | | Release date: | 2020-09-16 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (2.98 Å) | | Cite: | The molecular basis of tight nuclear tethering and inactivation of cGAS.
Nature, 587, 2020
|
|
8U9G
 
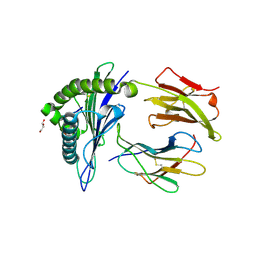 | | Human Class I MHC HLA-A2 bound to sorting nexin 24 (127-135) neoantigen KLSHQLVLL | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Beta-2-microglobulin, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Arbuiso, A, Weiss, L.I, Brambley, C.A, Ma, J, Keller, G.L.J, Ayres, C.M, Baker, B.M. | | Deposit date: | 2023-09-19 | | Release date: | 2024-01-10 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.87 Å) | | Cite: | Accurate modeling of peptide-MHC structures with AlphaFold.
Structure, 32, 2024
|
|
6WCZ
 
 | | CryoEM structure of full-length ZIKV NS5-hSTAT2 complex | | Descriptor: | Non-structural protein 5, Signal transducer and activator of transcription 2, ZINC ION | | Authors: | Boxiao, W, Stephanie, T, Kang, Z, Maria, T.S, Jian, F, Jiuwei, L, Linfeng, G, Wendan, R, Yanxiang, C, Ethan, C.V, HeaJin, H, Matthew, J.E, Sean, E.O, Adolfo, G.S, Hong, Z, Rong, H, Jikui, S. | | Deposit date: | 2020-03-31 | | Release date: | 2020-07-08 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structural basis for STAT2 suppression by flavivirus NS5.
Nat.Struct.Mol.Biol., 27, 2020
|
|
8TBV
 
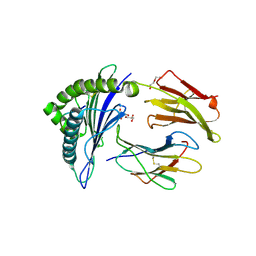 | | Human Class I MHC HLA-A2 in complex with sorting nexin 24 (127-135) neoantigen KLSHQLVLL | | Descriptor: | Beta-2-microglobulin, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Arbuiso, A, Weiss, L.I, Brambley, C.A, Ma, J, Keller, G.L.J, Ayres, C.M, Baker, B.M. | | Deposit date: | 2023-06-29 | | Release date: | 2024-01-17 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.64 Å) | | Cite: | Accurate modeling of peptide-MHC structures with AlphaFold.
Structure, 32, 2024
|
|
8TBW
 
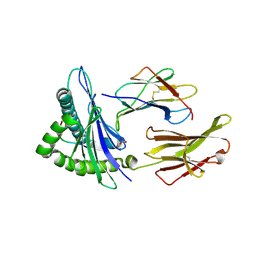 | | Human Class I MHC HLA-A2 in complex with sorting nexin 24 (127-135) peptide KLSHQPVLL | | Descriptor: | Beta-2-microglobulin, HLA-A*02:01 alpha chain, NITRATE ION, ... | | Authors: | Arbuiso, A, Weiss, L.I, Brambley, C.A, Ma, J, Keller, G.L.J, Ayres, C.M, Baker, B.M. | | Deposit date: | 2023-06-29 | | Release date: | 2024-01-10 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.081 Å) | | Cite: | Accurate modeling of peptide-MHC structures with AlphaFold.
Structure, 32, 2024
|
|
8I6Q
 
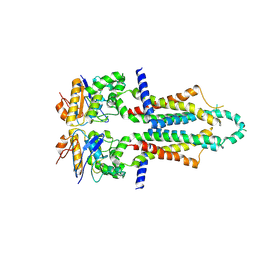 | |
4BLF
 
 | | Variable internal flexibility characterizes the helical capsid formed by Agrobacterium VirE2 protein on single-stranded DNA. | | Descriptor: | SINGLE-STRAND DNA-BINDING PROTEIN | | Authors: | Bharat, T.A.M, Zbaida, D, Eisenstein, M, Frankenstein, Z, Mehlman, T, Weiner, L, Sorzano, C.O.S, Barak, Y, Albeck, S, Briggs, J.A.G, Wolf, S.G, Elbaum, M. | | Deposit date: | 2013-05-02 | | Release date: | 2013-06-26 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (20 Å) | | Cite: | Variable Internal Flexibility Characterizes the Helical Capsid Formed by Agrobacterium Vire2 Protein on Single-Stranded DNA.
Structure, 21, 2013
|
|
2F6H
 
 | |
7CJF
 
 | | Crystal structure of SARS-CoV-2 RBD in complex with a neutralizing antibody Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike protein S1, antibody heavy chain, ... | | Authors: | Guo, Y, Li, X, Zhang, G, Fu, D, Schweizer, L, Zhang, H, Rao, Z. | | Deposit date: | 2020-07-10 | | Release date: | 2020-11-11 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.108 Å) | | Cite: | A SARS-CoV-2 neutralizing antibody with extensive Spike binding coverage and modified for optimal therapeutic outcomes.
Nat Commun, 12, 2021
|
|
3FMQ
 
 | | Crystal structure of an Encephalitozoon cuniculi methionine aminopeptidase type 2 with angiogenesis inhibitor fumagillin bound | | Descriptor: | FE (III) ION, FUMAGILLIN, Methionine aminopeptidase 2, ... | | Authors: | Alvarado, J.J, Russell, M, Zhang, A, Adams, J, Toro, R, Burley, S.K, Weiss, L.M, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-12-22 | | Release date: | 2009-01-13 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of a microsporidian methionine aminopeptidase type 2 complexed with fumagillin and TNP-470.
Mol.Biochem.Parasitol., 168, 2009
|
|
3FMR
 
 | | Crystal structure of an Encephalitozoon cuniculi methionine aminopeptidase type 2 with angiogenesis inhibitor TNP470 bound | | Descriptor: | (1R,2S,3S,4R)-4-hydroxy-2-methoxy-4-methyl-3-[(2R,3R)-2-methyl-3-(3-methylbut-2-en-1-yl)oxiran-2-yl]cyclohexyl (chloroacetyl)carbamate, FE (III) ION, Methionine aminopeptidase 2, ... | | Authors: | Alvarado, J.J, Russell, M, Zhang, A, Adams, J, Toro, R, Burley, S.K, Weiss, L.M, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-12-22 | | Release date: | 2009-01-13 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.89 Å) | | Cite: | Structure of a microsporidian methionine aminopeptidase type 2 complexed with fumagillin and TNP-470.
Mol.Biochem.Parasitol., 168, 2009
|
|
3FM3
 
 | | Crystal structure of an Encephalitozoon cuniculi methionine aminopeptidase type 2 | | Descriptor: | FE (III) ION, Methionine aminopeptidase 2, SULFATE ION | | Authors: | Alvarado, J.J, Russell, M, Zhang, A, Adams, J, Toro, R, Burley, S.K, Weiss, L.M, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-12-19 | | Release date: | 2009-01-13 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Structure of a microsporidian methionine aminopeptidase type 2 complexed with fumagillin and TNP-470.
Mol.Biochem.Parasitol., 168, 2009
|
|
2J4F
 
 | | Torpedo acetylcholinesterase - Hg heavy-atom derivative | | Descriptor: | ACETYLCHOLINESTERASE, MERCURY (II) ION | | Authors: | Kreimer, D.I, Dolginova, E.A, Raves, M, Sussman, J.L, Silman, I, Weiner, L. | | Deposit date: | 2006-08-30 | | Release date: | 2006-09-05 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | A Metastable State of Torpedo Californica Acetylcholinesterase Generated by Modification with Organomercurials
Biochemistry, 33, 1994
|
|
