4MLG
 
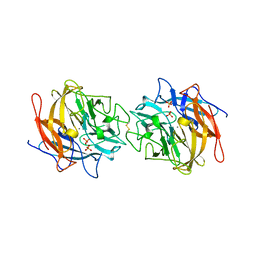 | | Structure of RS223-Beta-xylosidase | | Descriptor: | Beta-xylosidase, CALCIUM ION, SULFATE ION | | Authors: | Jordan, D, Braker, J, Wagschal, K, Lee, C, Dubrovska, I, Anderson, S, Wawrzak, Z. | | Deposit date: | 2013-09-06 | | Release date: | 2014-09-17 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure of RS223-Beta-xylosidase
To be Published
|
|
7UWP
 
 | | Detergent-bound CYP51 from Acanthamoeba castellanii | | Descriptor: | DODECYL-BETA-D-MALTOSIDE, PROTOPORPHYRIN IX CONTAINING FE, sterol 14a-demethylase | | Authors: | Hargrove, T.Y, Wawrzak, Z, Lepesheva, G.I. | | Deposit date: | 2022-05-03 | | Release date: | 2023-04-12 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Identification of Potent and Selective Inhibitors of Acanthamoeba: Structural Insights into Sterol 14-Demethylase as a Key Drug Target
J.Med.Chem., 2024
|
|
4MHB
 
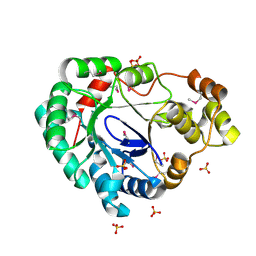 | | Structure of a putative reductase from Yersinia pestis | | Descriptor: | Putative aldo/keto reductase, SULFATE ION | | Authors: | Anderson, S.M, Wawrzak, Z, Kudritska, M, Kwon, K, Rembert, P, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-08-29 | | Release date: | 2013-10-16 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structure of a putative reductase from Yersinia pestis
To be Published
|
|
4W8K
 
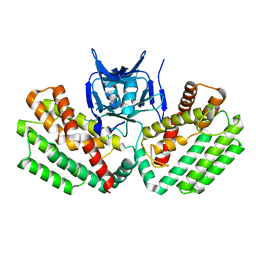 | | Crystal structure of a putative Cas1 enzyme from Vibrio phage ICP1 | | Descriptor: | Cas1 protein, POTASSIUM ION | | Authors: | Stogios, P.J, Wawrzak, Z, Onopriyeno, O, Yim, V, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2014-08-25 | | Release date: | 2014-09-17 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | To be published
To Be Published
|
|
6UEZ
 
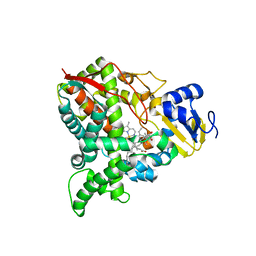 | |
6VBB
 
 | | 2.60 Angstrom Resolution Crystal Structure of Peptidase S41 from Acinetobacter baumannii | | Descriptor: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, Peptidase S41, ... | | Authors: | Minasov, G, Wawrzak, Z, Shuvalova, L, Kiryukhina, O, Dubrovska, I, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2019-12-18 | | Release date: | 2019-12-25 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | 2.60 Angstrom Resolution Crystal Structure of Peptidase S41 from Acinetobacter baumannii
To Be Published
|
|
1VRO
 
 | | Selenium-Assisted Nucleic Acid Crystallography: Use of Phosphoroselenoates for MAD Phasing of a DNA Structure | | Descriptor: | 5'-D(*CP*(GMS)P*CP*GP*CP*G)-3', MAGNESIUM ION, SPERMINE | | Authors: | Wilds, C.J, Pattanayek, R, Pan, C, Wawrzak, Z, Egli, M. | | Deposit date: | 2005-04-14 | | Release date: | 2005-04-19 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Selenium-Assisted Nucleic Acid Crystallography: Use of Phosphoroselenoates for MAD Phasing of a DNA Structure
J.Am.Chem.Soc., 124, 2002
|
|
1W5U
 
 | | GRAMICIDIN D FROM BACILLUS BREVIS (ETHANOL SOLVATE) | | Descriptor: | CHLORIDE ION, ETHANOL, GRAMICIDIN D, ... | | Authors: | Glowka, M.L, Olczak, A, Bojarska, J, Szczesio, M, Duax, W.L, Burkhart, B.M, Pangborn, W.A, Langs, D.A, Wawrzak, Z. | | Deposit date: | 2004-08-10 | | Release date: | 2005-09-15 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.14 Å) | | Cite: | Structure of Gramicidin D-Rbcl Complex at Atomic Resolution from Low-Temperature Synchrotron Data: Interactions of Double-Stranded Gramicidin Channel Contents and Cations with Channel Wall
Acta Crystallogr.,Sect.D, 61, 2005
|
|
8SBI
 
 | |
2IZQ
 
 | | Gramicidin D complex with KI | | Descriptor: | GRAMICIDIN D, IODIDE ION, METHANOL, ... | | Authors: | Olczak, A, Glowka, M.L, Szczesio, M, Bojarska, J, Duax, W.L, Burkhart, B.M, Wawrzak, Z. | | Deposit date: | 2006-07-26 | | Release date: | 2007-01-23 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (0.8 Å) | | Cite: | Nonstoichiometric Complex of Gramicidin D with Ki at 0.80 A Resolution.
Acta Crystallogr.,Sect.D, 63, 2007
|
|
7LO9
 
 | | RNA dodecamer containing a GNA A residue | | Descriptor: | Chains: A,B,C,D | | Authors: | Harp, J.M, Wawrzak, Z, Egli, M. | | Deposit date: | 2021-02-09 | | Release date: | 2021-12-22 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Overcoming GNA/RNA base-pairing limitations using isonucleotides improves the pharmacodynamic activity of ESC+ GalNAc-siRNAs.
Nucleic Acids Res., 49, 2021
|
|
4UYM
 
 | |
4Z8Z
 
 | | Crystal structure of the hypothetical protein from Ruminiclostridium thermocellum ATCC 27405 | | Descriptor: | Uncharacterized protein | | Authors: | Filippova, E.V, Wawrzak, Z, Kiryukhina, O, Endres, M, Joachimiak, J, Anderson, W.F, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2015-04-09 | | Release date: | 2015-05-06 | | Last modified: | 2019-12-11 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Crystal structure of the hypothetical protein from Ruminiclostridium thermocellum ATCC 27405
To Be Published
|
|
4ZV9
 
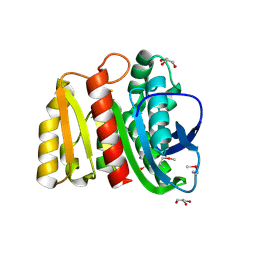 | | 2.00 Angstrom resolution crystal structure of an uncharacterized protein from Escherichia coli O157:H7 str. Sakai | | Descriptor: | DI(HYDROXYETHYL)ETHER, GLYCEROL, PHOSPHATE ION, ... | | Authors: | Halavaty, A.S, Wawrzak, Z, Filippova, E.V, Kiryukhina, O, Endres, M, Joachimiak, A, Anderson, W.F, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2015-05-18 | | Release date: | 2015-06-17 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | 2.00 Angstrom resolution crystal structure of an uncharacterized protein from Escherichia coli O157:H7 str. Sakai
To Be Published
|
|
3ETF
 
 | | Crystal structure of a putative succinate-semialdehyde dehydrogenase from salmonella typhimurium lt2 | | Descriptor: | Putative succinate-semialdehyde dehydrogenase | | Authors: | Brunzelle, J.S, Evdokimova, E, Kudritska, M, Wawrzak, Z, Anderson, W.F, Savchenk, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2008-10-07 | | Release date: | 2008-11-04 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure and activity of the NAD(P)(+) -dependent succinate semialdehyde dehydrogenase YneI from Salmonella typhimurium.
Proteins, 81, 2013
|
|
1TW7
 
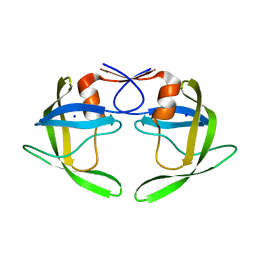 | | Wide Open 1.3A Structure of a Multi-drug Resistant HIV-1 Protease Represents a Novel Drug Target | | Descriptor: | SODIUM ION, protease | | Authors: | Martin, P, Vickrey, J.F, Proteasa, G, Jimenez, Y.L, Wawrzak, Z, Winters, M.A, Merigan, T.C, Kovari, L.C. | | Deposit date: | 2004-06-30 | | Release date: | 2005-07-19 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Wide Open 1.3A Structure of a Multi-drug Resistant HIV-1 Protease Represents a Novel Drug Target
Structure, 13, 2005
|
|
3GEU
 
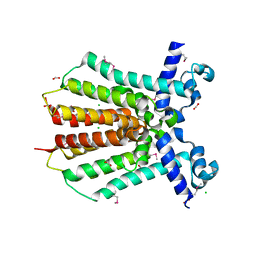 | | Crystal Structure of IcaR from Staphylococcus aureus, a member of the tetracycline repressor protein family | | Descriptor: | CHLORIDE ION, FORMIC ACID, Intercellular adhesion protein R, ... | | Authors: | Anderson, S.M, Brunzelle, J.S, Wawrzak, Z, Skarina, T, Papazisi, L, Anderson, W.F, Savchenko, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2009-02-26 | | Release date: | 2009-03-10 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure of IcaR from Staphylococcus aureus, a member of the tetracycline repressor protein family
To be Published
|
|
6AON
 
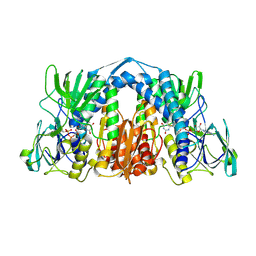 | | 1.72 Angstrom Resolution Crystal Structure of 2-Oxoglutarate Dehydrogenase Complex Subunit Dihydrolipoamide Dehydrogenase from Bordetella pertussis in Complex with FAD | | Descriptor: | CALCIUM ION, Dihydrolipoyl dehydrogenase, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Minasov, G, Wawrzak, Z, Skarina, T, McChesney, C, Grimshaw, S, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2017-08-16 | | Release date: | 2017-08-23 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | 1.72 Angstrom Resolution Crystal Structure of 2-Oxoglutarate Dehydrogenase Complex Subunit Dihydrolipoamide Dehydrogenase from Bordetella pertussis in Complex with FAD
To Be Published
|
|
6AOO
 
 | | 2.15 Angstrom Resolution Crystal Structure of Malate Dehydrogenase from Haemophilus influenzae | | Descriptor: | Malate dehydrogenase, SULFATE ION | | Authors: | Minasov, G, Wawrzak, Z, Skarina, T, Grimshaw, S, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2017-08-16 | | Release date: | 2017-08-23 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | 2.15 Angstrom Resolution Crystal Structure of Malate Dehydrogenase from Haemophilus influenzae.
To Be Published
|
|
6AZI
 
 | | 1.75 Angstrom Resolution Crystal Structure of D-alanyl-D-alanine Endopeptidase from Enterobacter cloacae in Complex with Covalently Bound Boronic Acid | | Descriptor: | BORATE ION, D-alanyl-D-alanine endopeptidase | | Authors: | Minasov, G, Wawrzak, Z, Shuvalova, L, Kiryukhina, O, Dubrovska, I, Grimshaw, S, Kwon, K, Anderson, W.F, Satchell, K.J.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2017-09-11 | | Release date: | 2017-10-04 | | Last modified: | 2023-05-31 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | 1.75 Angstrom Resolution Crystal Structure of D-alanyl-D-alanine Endopeptidase from Enterobacter cloacae in Complex with Covalently Bound Boronic Acid.
To be Published
|
|
7RTQ
 
 | | Sterol 14alpha demethylase (CYP51) from Naegleria fowleri in complex with an inhibitor R)-N-(1-(3,4'-difluorobiphenyl-4-yl)-2-(1H-imidazol-1-yl)ethyl)-4-(5-phenyl-1,3,4-oxadiazol-2-yl)benzamide | | Descriptor: | N-[(1R)-2-(1H-imidazol-1-yl)-1-(3,4',5-trifluoro[1,1'-biphenyl]-4-yl)ethyl]-4-(5-phenyl-1,3,4-oxadiazol-2-yl)benzamide, PROTOPORPHYRIN IX CONTAINING FE, Protein CYP51 | | Authors: | Lepesheva, G.I, Hargrove, T.Y, Wawrzak, Z. | | Deposit date: | 2021-08-13 | | Release date: | 2021-11-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Relaxed Substrate Requirements of Sterol 14 alpha-Demethylase from Naegleria fowleri Are Accompanied by Resistance to Inhibition.
J.Med.Chem., 64, 2021
|
|
1N1O
 
 | | Crystal Structure of a B-form DNA Duplex Containing (L)-alpha-threofuranosyl (3'-2') Nucleosides: A Four-Carbon Sugar is Easily Accommodated into the Backbone of DNA | | Descriptor: | 5'-D(*CP*GP*CP*GP*AP*AP*(TFT)P*TP*CP*GP*CP*G)-3', MAGNESIUM ION | | Authors: | Wilds, C.J, Wawrzak, Z, Krishnamurthy, R, Eschenmoser, A, Egli, M. | | Deposit date: | 2002-10-18 | | Release date: | 2002-11-15 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Crystal Structure of a B-Form DNA Duplex Containing (L)-alpha-Threofuranosyl (3'-->2') Nucleosides: A
Four-Carbon Sugar Is Easily Accommodated into the Backbone of DNA
J.Am.Chem.Soc., 124, 2002
|
|
5KVR
 
 | | X-Ray Crystal Structure of a Fragment (1-75) of a Transcriptional Regulator PdhR from Escherichia coli CFT073 | | Descriptor: | Pyruvate dehydrogenase complex repressor | | Authors: | Brunzelle, J.S, Wawrzak, Z, Sandoval, J, Skarina, T, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-07-15 | | Release date: | 2016-11-02 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | X-Ray Crystal Structure of a Fragment (1-75) of a Transcriptional Regulator PdhR from Escherichia coli CFT073
To Be Published
|
|
7SNM
 
 | |
1PNV
 
 | | Crystal Structure of TDP-epi-Vancosaminyltransferase GtfA in complexes with TDP and Vancomycin | | Descriptor: | GLYCOSYLTRANSFERASE GTFA, THYMIDINE-5'-DIPHOSPHATE, VANCOMYCIN, ... | | Authors: | Mulichak, A.M, Losey, H.C, Lu, W, Wawrzak, Z, Walsh, C.T, Garavito, R.M. | | Deposit date: | 2003-06-13 | | Release date: | 2003-08-12 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of the Tdp-Epi-Vancosaminyltransferase Gtfa from the Chloroeremomycin Biosynthetic Pathway.
Proc.Natl.Acad.Sci.USA, 100, 2003
|
|
