5KJT
 
 | | Crystal structure of Arabidopsis thaliana HCT in complex with p-coumaroyl-CoA | | Descriptor: | Shikimate O-hydroxycinnamoyltransferase, p-coumaroyl-CoA | | Authors: | Levsh, O, Chiang, Y.C, Tung, C.F, Noel, J.P, Wang, Y, Weng, J.K. | | Deposit date: | 2016-06-20 | | Release date: | 2016-11-02 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Dynamic Conformational States Dictate Selectivity toward the Native Substrate in a Substrate-Permissive Acyltransferase.
Biochemistry, 55, 2016
|
|
5KJW
 
 | | Crystal structure of Coleus blumei HCT in complex with 3-hydroxyacetophenone | | Descriptor: | 1-(3-hydroxyphenyl)ethanone, Hydroxycinnamoyl transferase | | Authors: | Levsh, O, Chiang, Y.C, Tung, C.F, Noel, J.P, Wang, Y, Weng, J.K. | | Deposit date: | 2016-06-20 | | Release date: | 2016-11-02 | | Last modified: | 2019-11-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Dynamic Conformational States Dictate Selectivity toward the Native Substrate in a Substrate-Permissive Acyltransferase.
Biochemistry, 55, 2016
|
|
5KJU
 
 | | Crystal structure of Arabidopsis thaliana HCT in complex with p-coumaroylshikimate | | Descriptor: | (3~{R},4~{S},5~{R})-3-[(~{E})-3-(4-hydroxyphenyl)prop-2-enoyl]oxy-4,5-bis(oxidanyl)cyclohexene-1-carboxylic acid, Shikimate O-hydroxycinnamoyltransferase | | Authors: | Levsh, O, Chiang, Y.C, Tung, C.F, Noel, J.P, Wang, Y, Weng, J.K. | | Deposit date: | 2016-06-20 | | Release date: | 2016-11-02 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | Dynamic Conformational States Dictate Selectivity toward the Native Substrate in a Substrate-Permissive Acyltransferase.
Biochemistry, 55, 2016
|
|
5VEW
 
 | | Structure of the human GLP-1 receptor complex with PF-06372222 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Glucagon-like peptide 1 receptor,Endolysin chimera, N-{4-[(R)-(3,3-dimethylcyclobutyl)({6-[4-(trifluoromethyl)-1H-imidazol-1-yl]pyridin-3-yl}amino)methyl]benzene-1-carbonyl}-beta-alanine, ... | | Authors: | Song, G, Yang, D, Wang, Y, Graaf, C.D, Zhou, Q, Jiang, S, Liu, K, Cai, X, Dai, A, Lin, G, Liu, D, Wu, F, Wu, Y, Zhao, S, Ye, L, Han, G.W, Lau, J, Wu, B, Hanson, M.A, Liu, Z.-J, Wang, M.-W, Stevens, R.C. | | Deposit date: | 2017-04-05 | | Release date: | 2017-05-24 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Human GLP-1 receptor transmembrane domain structure in complex with allosteric modulators.
Nature, 546, 2017
|
|
7JVQ
 
 | | Cryo-EM structure of apomorphine-bound dopamine receptor 1 in complex with Gs protein | | Descriptor: | (6aR)-6-methyl-5,6,6a,7-tetrahydro-4H-dibenzo[de,g]quinoline-10,11-diol, CHOLESTEROL, D(1A) dopamine receptor, ... | | Authors: | Zhuang, Y, Xu, P, Mao, C, Wang, L, Krumm, B, Zhou, X.E, Huang, S, Liu, H, Cheng, X, Huang, X.-P, Sheng, D.-D, Xu, T, Liu, Y.-F, Wang, Y, Guo, J, Jiang, Y, Jiang, H, Melcher, K, Roth, B.L, Zhang, Y, Zhang, C, Xu, H.E. | | Deposit date: | 2020-08-22 | | Release date: | 2021-02-24 | | Last modified: | 2021-03-03 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural insights into the human D1 and D2 dopamine receptor signaling complexes.
Cell, 184, 2021
|
|
7JVP
 
 | | Cryo-EM structure of SKF-83959-bound dopamine receptor 1 in complex with Gs protein | | Descriptor: | (1R)-6-chloro-3-methyl-1-(3-methylphenyl)-2,3,4,5-tetrahydro-1H-3-benzazepine-7,8-diol, CHOLESTEROL, D(1A) dopamine receptor, ... | | Authors: | Zhuang, Y, Xu, P, Mao, C, Wang, L, Krumm, B, Zhou, X.E, Huang, S, Liu, H, Cheng, X, Huang, X.-P, Sheng, D.-D, Xu, T, Liu, Y.-F, Wang, Y, Guo, J, Jiang, Y, Jiang, H, Melcher, K, Roth, B.L, Zhang, Y, Zhang, C, Xu, H.E. | | Deposit date: | 2020-08-22 | | Release date: | 2021-02-24 | | Last modified: | 2021-03-03 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural insights into the human D1 and D2 dopamine receptor signaling complexes.
Cell, 184, 2021
|
|
7JV5
 
 | | Cryo-EM structure of SKF-81297-bound dopamine receptor 1 in complex with Gs protein | | Descriptor: | (1R)-6-chloro-1-phenyl-2,3,4,5-tetrahydro-1H-3-benzazepine-7,8-diol, CHOLESTEROL, D(1A) dopamine receptor, ... | | Authors: | Zhuang, Y, Xu, P, Mao, C, Wang, L, Krumm, B, Zhou, X.E, Huang, S, Liu, H, Cheng, X, Huang, X.-P, Sheng, D.-D, Xu, T, Liu, Y.-F, Wang, Y, Guo, J, Jiang, Y, Jiang, H, Melcher, K, Roth, B.L, Zhang, Y, Zhang, C, Xu, H.E. | | Deposit date: | 2020-08-20 | | Release date: | 2021-02-24 | | Last modified: | 2021-03-03 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural insights into the human D1 and D2 dopamine receptor signaling complexes.
Cell, 184, 2021
|
|
7JVR
 
 | | Cryo-EM structure of Bromocriptine-bound dopamine receptor 2 in complex with Gi protein | | Descriptor: | Antibody fragment ScFv16, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Zhuang, Y, Xu, P, Mao, C, Wang, L, Krumm, B, Zhou, X.E, Huang, S, Liu, H, Cheng, X, Huang, X.-P, Sheng, D.-D, Xu, T, Liu, Y.-F, Wang, Y, Guo, J, Jiang, Y, Jiang, H, Melcher, K, Roth, B.L, Zhang, Y, Zhang, C, Xu, H.E. | | Deposit date: | 2020-08-22 | | Release date: | 2021-02-24 | | Last modified: | 2021-03-31 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structural insights into the human D1 and D2 dopamine receptor signaling complexes.
Cell, 184, 2021
|
|
5H1M
 
 | | Crystal structure of WD40 repeat domains of Gemin5 in complex with M7G | | Descriptor: | 7N-METHYL-8-HYDROGUANOSINE-5'-DIPHOSPHATE, Gem-associated protein 5 | | Authors: | Jin, W, Wang, Y, Liu, C.P, Yang, N, Jin, M, Cong, Y, Wang, M, Xu, R.M. | | Deposit date: | 2016-10-10 | | Release date: | 2016-11-23 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.492 Å) | | Cite: | Structural basis for snRNA recognition by the double-WD40 repeat domain of Gemin5
Genes Dev., 30, 2016
|
|
5XAF
 
 | | Crystal structure of tubulin-stathmin-TTL-Compound Z1 complex | | Descriptor: | (3S,4R)-4-(3-hydroxy-4-methoxyphenyl)-3-methyl-1-(3,4,5-trimethoxyphenyl)azetidin-2-one, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, ... | | Authors: | Zhang, H, Luo, C, Wang, Y. | | Deposit date: | 2017-03-12 | | Release date: | 2017-12-20 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.551 Å) | | Cite: | Design, synthesis, biological evaluation and cocrystal structures with tubulin of chiral beta-lactam bridged combretastatin A-4 analogues as potent antitumor agents
Eur J Med Chem, 144, 2017
|
|
3GO6
 
 | | Crystal Structure of M. tuberculosis ribokinase (Rv2436) in complex with ribose and AMP-PNP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, RIBOKINASE RBSK, ... | | Authors: | Masters, E.I, Sun, Y, Wang, Y, Parker, W.B, Li, R. | | Deposit date: | 2009-03-18 | | Release date: | 2010-03-31 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Crystal structure and biochemical characterization of M.tuberculosis ribokinase (Rv2436)
To be Published
|
|
6WWC
 
 | |
6WX2
 
 | |
6X2Q
 
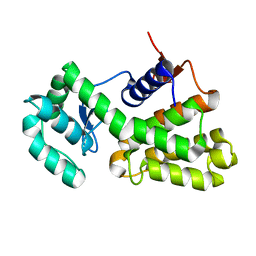 | | Complex of Gynuella sunshinyii GH46 chitosanase GsCsn46A with chitotetraose | | Descriptor: | 2-amino-2-deoxy-beta-D-glucopyranose, 2-amino-2-deoxy-beta-D-glucopyranose-(1-4)-2-amino-2-deoxy-beta-D-glucopyranose-(1-4)-2-amino-2-deoxy-beta-D-glucopyranose, Chitosanase | | Authors: | Qin, Z, Wang, Y. | | Deposit date: | 2020-05-20 | | Release date: | 2021-05-26 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Complex of Gynuella sunshinyii GH46 chitosanase GsCsn46A with chitotetraose
To Be Published
|
|
4XKL
 
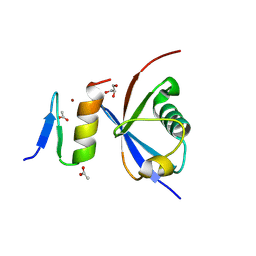 | | Crystal structure of NDP52 ZF2 in complex with mono-ubiquitin | | Descriptor: | ACETATE ION, Calcium-binding and coiled-coil domain-containing protein 2, GLYCEROL, ... | | Authors: | Xie, X, Li, F, Wang, Y, Lin, Z, Chen, X, Liu, J, Pan, L. | | Deposit date: | 2015-01-12 | | Release date: | 2015-11-11 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Molecular basis of ubiquitin recognition by the autophagy receptor CALCOCO2
Autophagy, 11, 2015
|
|
7VI7
 
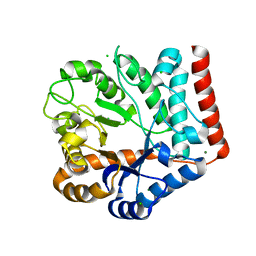 | | Crystal structure of GH3 beta-N-acetylhexosaminidase Amuc_2109 from Akkermansia muciniphila in complex with GlcNAc | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose, Beta-N-acetylhexosaminidase, CHLORIDE ION, ... | | Authors: | Qian, K, Yang, W, Chen, X, Wang, Y, Zhang, M, Wang, M. | | Deposit date: | 2021-09-26 | | Release date: | 2021-12-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Functional and structural characterization of a GH3 beta-N-acetylhexosaminidase from Akkermansia muciniphila involved in mucin degradation
Biochem.Biophys.Res.Commun., 589, 2022
|
|
7VI6
 
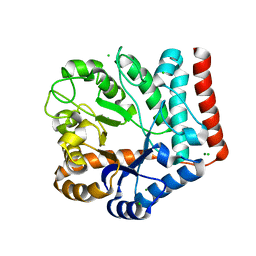 | | Crystal structure of GH3 beta-N-acetylhexosaminidase Amuc_2109 from Akkermansia muciniphila | | Descriptor: | Beta-N-acetylhexosaminidase, CHLORIDE ION, MAGNESIUM ION | | Authors: | Qian, K, Yang, W, Chen, X, Wang, Y, Zhang, M, Wang, M. | | Deposit date: | 2021-09-26 | | Release date: | 2021-12-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Functional and structural characterization of a GH3 beta-N-acetylhexosaminidase from Akkermansia muciniphila involved in mucin degradation
Biochem.Biophys.Res.Commun., 589, 2022
|
|
3GO7
 
 | | Crystal Structure of M. tuberculosis ribokinase (Rv2436) in complex with ribose | | Descriptor: | MAGNESIUM ION, RIBOKINASE RBSK, alpha-D-ribofuranose | | Authors: | Masters, E.I, Sun, Y, Wang, Y, Parker, W.B, Li, R. | | Deposit date: | 2009-03-18 | | Release date: | 2010-03-31 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure and biochemical characterization of M.tuberculosis ribokinase (Rv2436)
To be Published
|
|
3HHS
 
 | | Crystal Structure of Manduca sexta prophenoloxidase | | Descriptor: | COPPER (II) ION, Phenoloxidase subunit 1, Phenoloxidase subunit 2 | | Authors: | Li, Y, Wang, Y, Jiang, H, Deng, J. | | Deposit date: | 2009-05-17 | | Release date: | 2009-09-29 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Crystal structure of Manduca sexta prophenoloxidase provides insights into the mechanism of type 3 copper enzymes.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
7PEM
 
 | | Cryo-EM structure of phophorylated Drs2p-Cdc50p in a PS and ATP-bound E2P state | | Descriptor: | (2R)-1-{[(R)-hydroxy{[(1R,2R,3R,4R,5S,6R)-2,3,5,6-tetrahydroxy-4-(phosphonooxy)cyclohexyl]oxy}phosphoryl]oxy}-3-(octadecanoyloxy)propan-2-yl (5Z,8Z,11Z,14Z)-icosa-5,8,11,14-tetraenoate, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Timcenko, M, Wang, Y, Lyons, J.A, Nissen, P, Lindorff-Larsen, K. | | Deposit date: | 2021-08-10 | | Release date: | 2022-08-24 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Substrate Transport and Specificity in a Phospholipid Flippase
To Be Published
|
|
5GNF
 
 | | Crystal structure of anti-CRISPR protein AcrF3 | | Descriptor: | CALCIUM ION, Uncharacterized protein AcrF3 | | Authors: | Wang, J, Wang, Y. | | Deposit date: | 2016-07-20 | | Release date: | 2016-09-21 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | A CRISPR evolutionary arms race: structural insights into viral anti-CRISPR/Cas responses
Cell Res., 26, 2016
|
|
5GQH
 
 | | Cryo-EM structure of PaeCas3-AcrF3 complex | | Descriptor: | CRISPR-associated nuclease/helicase Cas3 subtype I-F/YPEST, anti-CRISPR protein 3 | | Authors: | Zhang, X, Ma, J, Wang, Y, Wang, J. | | Deposit date: | 2016-08-07 | | Release date: | 2016-09-21 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | A CRISPR evolutionary arms race: structural insights into viral anti-CRISPR/Cas responses
Cell Res., 26, 2016
|
|
8GXD
 
 | | L-LEUCINE DEHYDROGENASE FROM EXIGUOBACTERIUM SIBIRICUM | | Descriptor: | CALCIUM ION, GLYCEROL, Glu/Leu/Phe/Val dehydrogenase | | Authors: | Mu, X, Nie, Y, Wu, T, Wang, Y, Zhang, N, Yin, D, Xu, Y. | | Deposit date: | 2022-09-19 | | Release date: | 2023-04-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.02 Å) | | Cite: | Reshaping Substrate-Binding Pocket of Leucine Dehydrogenase for Bidirectionally Accessing Structurally Diverse Substrates
Acs Catalysis, 13, 2023
|
|
2YJY
 
 | | A specific and modular binding code for cytosine recognition in PUF domains | | Descriptor: | 5'-R(*AP*UP*UP*GP*CP*AP*UP*AP*UP*AP)-3', PUMILIO HOMOLOG 1 | | Authors: | Dong, S, Wang, Y, Cassidy-Amstutz, C, Lu, G, Qiu, C, Bigler, R, Jezyk, M, Li, C, Hall, T.M.T, Wang, Z. | | Deposit date: | 2011-05-24 | | Release date: | 2011-06-08 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.598 Å) | | Cite: | Specific and Modular Binding Code for Cytosine Recognition in Pumilio/Fbf (Puf) RNA-Binding Domains.
J.Biol.Chem., 286, 2011
|
|
7VFV
 
 | | Human N-type voltage gated calcium channel CaV2.2-alpha2/delta1-beta1 complex, bound to PD173212 | | Descriptor: | (2~{S})-~{N}-[(2~{S})-1-(~{tert}-butylamino)-1-oxidanylidene-3-(4-phenylmethoxyphenyl)propan-2-yl]-2-[(4-~{tert}-butylphenyl)methyl-methyl-amino]-4-methyl-pentanamide, 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Dong, Y, Gao, Y, Wang, Y, Zhao, Y. | | Deposit date: | 2021-09-13 | | Release date: | 2021-11-03 | | Last modified: | 2022-02-16 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Closed-state inactivation and pore-blocker modulation mechanisms of human Ca V 2.2.
Cell Rep, 37, 2021
|
|
