7WRV
 
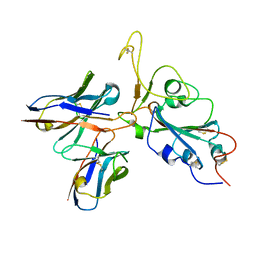 | | The interface of JMB2002 Fab binds to SARS-CoV-2 Omicron Variant S | | Descriptor: | JMB2002 Fab heavy chain, JMB2002 Fab light chain, Spike glycoprotein | | Authors: | Yin, W, Xu, Y, Xu, P, Cao, X, Wu, C, Gu, C, He, X, Wang, X, Huang, S, Yuan, Q, Wu, K, Hu, W, Huang, Z, Liu, J, Wang, Z, Jia, F, Xia, K, Liu, P, Wang, X, Song, B, Zheng, J, Jiang, H, Cheng, X, Jiang, Y, Deng, S.J, Xu, H.E. | | Deposit date: | 2022-01-27 | | Release date: | 2022-03-23 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (2.47 Å) | | Cite: | Structures of the Omicron spike trimer with ACE2 and an anti-Omicron antibody.
Science, 375, 2022
|
|
6ITM
 
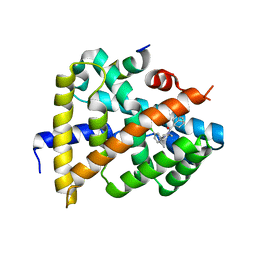 | | Crystal structure of FXR in complex with agonist XJ034 | | Descriptor: | 1-adamantyl-[4-(5-chloranyl-2-methyl-phenyl)piperazin-1-yl]methanone, Bile acid receptor, HD3 Peptide from Nuclear receptor coactivator 1 | | Authors: | Zhang, H, Wang, Z. | | Deposit date: | 2018-11-23 | | Release date: | 2019-11-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Pose Filter-Based Ensemble Learning Enables Discovery of Orally Active, Nonsteroidal Farnesoid X Receptor Agonists.
J.Chem.Inf.Model., 60, 2020
|
|
1N5U
 
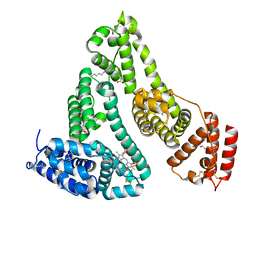 | | X-RAY STUDY OF HUMAN SERUM ALBUMIN COMPLEXED WITH HEME | | Descriptor: | MYRISTIC ACID, PROTOPORPHYRIN IX CONTAINING FE, SERUM ALBUMIN | | Authors: | Wardell, M, Wang, Z, Ho, J.X, Robert, J, Ruker, F, Ruble, J, Carter, D.C. | | Deposit date: | 2002-11-07 | | Release date: | 2003-06-24 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The Atomic Structure of Human Methemalbumin at 1.9 A
Biochem.Biophys.Res.Commun., 291, 2002
|
|
6UWD
 
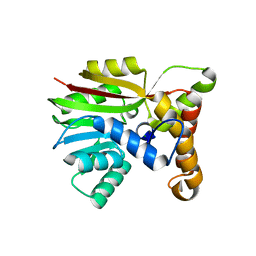 | | Crystal structure of Apo AtmM | | Descriptor: | ACETATE ION, D-glucose O-methyltransferase, MAGNESIUM ION | | Authors: | Alvarado, S.K, Wang, Z, Miller, M.D, Thorson, J.S, Phillips Jr, G.N. | | Deposit date: | 2019-11-05 | | Release date: | 2020-11-25 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Structure of Apo AtmM
To Be Published
|
|
5YKI
 
 | | Crystal structure of the engineered nine-repeat PUF domain in complex with cognate 9nt-RNA | | Descriptor: | Pumilio homolog 1, RNA (5'-R(*UP*GP*UP*UP*GP*UP*AP*UP*A)-3') | | Authors: | Zhao, Y.Y, Wang, J, Li, H.T, Wang, Z.X, Wu, J.W. | | Deposit date: | 2017-10-14 | | Release date: | 2018-03-14 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Expanding RNA binding specificity and affinity of engineered PUF domains.
Nucleic Acids Res., 46, 2018
|
|
3EQP
 
 | | Crystal Structure of Ack1 with compound T95 | | Descriptor: | Activated CDC42 kinase 1, CHLORIDE ION, N-(2,6-dimethylphenyl)-4-(2-ethoxyphenoxy)-2-({4-[4-(2-hydroxyethyl)piperazin-1-yl]phenyl}amino)pyrimidine-5-carboxamide | | Authors: | Liu, J, Wang, Z, Walker, N.P.C. | | Deposit date: | 2008-10-01 | | Release date: | 2008-12-02 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Identification and optimization of N3,N6-diaryl-1H-pyrazolo[3,4-d]pyrimidine-3,6-diamines as a novel class of ACK1 inhibitors.
Bioorg.Med.Chem.Lett., 18, 2008
|
|
5YKH
 
 | | Crystal structure of the engineered nine-repeat PUF domain | | Descriptor: | PHOSPHATE ION, Pumilio homolog 1 | | Authors: | Zhao, Y.Y, Wang, J, Li, H.T, Wang, Z.X, Wu, J.W. | | Deposit date: | 2017-10-14 | | Release date: | 2018-03-14 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.457 Å) | | Cite: | Expanding RNA binding specificity and affinity of engineered PUF domains.
Nucleic Acids Res., 46, 2018
|
|
7WPA
 
 | | SARS-CoV-2 Omicron Variant SPIKE trimer complexed with ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, ... | | Authors: | Yin, W, Xu, Y, Xu, P, Cao, X, Wu, C, Gu, C, He, X, Wang, X, Huang, S, Yuan, Q, Wu, K, Hu, W, Huang, Z, Liu, J, Wang, Z, Jia, F, Xia, K, Liu, P, Wang, X, Song, B, Zheng, J, Jiang, H, Cheng, X, Jiang, Y, Deng, S, Xu, E. | | Deposit date: | 2022-01-23 | | Release date: | 2022-03-09 | | Last modified: | 2022-03-16 | | Method: | ELECTRON MICROSCOPY (2.77 Å) | | Cite: | Structures of the Omicron spike trimer with ACE2 and an anti-Omicron antibody.
Science, 375, 2022
|
|
7WPD
 
 | | SARS-CoV-2 Omicron Variant S Trimer complexed with one JMB2002 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Anti-Fab nanobody, ... | | Authors: | Yin, W, Xu, Y, Xu, P, Cao, X, Wu, C, Gu, C, He, X, Wang, X, Huang, S, Yuan, Q, Wu, K, Hu, W, Huang, Z, Liu, J, Wang, Z, Jia, F, Xia, K, Liu, P, Wang, X, Song, B, Zheng, J, Jiang, H, Cheng, X, Jiang, Y, Deng, S.J, Xu, H.E. | | Deposit date: | 2022-01-23 | | Release date: | 2022-03-16 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.18 Å) | | Cite: | Structures of the Omicron spike trimer with ACE2 and an anti-Omicron antibody.
Science, 375, 2022
|
|
7WPF
 
 | | SARS-CoV-2 Omicron Variant S Trimer complexed with three JMB2002 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Anti-Fab nanobody, ... | | Authors: | Yin, W, Xu, Y, Xu, P, Cao, X, Wu, C, Gu, C, He, X, Wang, X, Huang, S, Yuan, Q, Wu, K, Hu, W, Huang, Z, Liu, J, Wang, Z, Jia, F, Xia, K, Liu, P, Wang, X, Song, B, Zheng, J, Jiang, H, Cheng, X, Jiang, Y, Deng, S.J, Xu, H.E. | | Deposit date: | 2022-01-23 | | Release date: | 2022-03-16 | | Method: | ELECTRON MICROSCOPY (2.92 Å) | | Cite: | Structures of the Omicron spike trimer with ACE2 and an anti-Omicron antibody.
Science, 375, 2022
|
|
5X6G
 
 | | Crystal Structure of SMAD5-MH1/palindromic SBE DNA complex | | Descriptor: | DNA (5'-D(P*AP*TP*CP*AP*GP*TP*CP*TP*AP*GP*AP*CP*AP*TP*A)-3'), DNA (5'-D(P*GP*TP*AP*TP*GP*TP*CP*TP*AP*GP*AP*CP*TP*GP*A)-3'), Mothers against decapentaplegic homolog 5, ... | | Authors: | Chai, N, Wang, J, Wang, Z.X, Wu, J.W. | | Deposit date: | 2017-02-21 | | Release date: | 2017-03-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Structural basis for the Smad5 MH1 domain to recognize different DNA sequences.
Nucleic Acids Res., 43, 2015
|
|
5X6M
 
 | | Crystal Structure of SMAD5-MH1 in complex with a composite DNA sequence | | Descriptor: | DNA (5'-D(P*AP*TP*CP*AP*GP*AP*CP*TP*GP*CP*CP*GP*GP*CP*AP*GP*TP*CP*TP*AP*TP*A)-3'), DNA (5'-D(P*TP*TP*AP*TP*AP*GP*AP*CP*TP*GP*CP*CP*GP*GP*CP*AP*GP*TP*CP*TP*GP*A)-3'), Mothers against decapentaplegic homolog 5, ... | | Authors: | Chai, N, Wang, J, Wang, Z.X, Wu, J.W. | | Deposit date: | 2017-02-22 | | Release date: | 2017-03-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural basis for the Smad5 MH1 domain to recognize different DNA sequences.
Nucleic Acids Res., 43, 2015
|
|
6O3V
 
 | | Crystal structure for RVA-VP3 | | Descriptor: | 1,2-ETHANEDIOL, GUANOSINE-5'-MONOPHOSPHATE, Protein VP3, ... | | Authors: | Kumar, D, Yu, X, Wang, Z, Hu, L, Prasad, V. | | Deposit date: | 2019-02-27 | | Release date: | 2020-03-04 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | 2.7 angstrom cryo-EM structure of rotavirus core protein VP3, a unique capping machine with a helicase activity.
Sci Adv, 6, 2020
|
|
7WPE
 
 | | SARS-CoV-2 Omicron Variant S Trimer complexed with two JMB2002 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Anti-Fab nanobody, ... | | Authors: | Yin, W, Xu, Y, Xu, P, Cao, X, Wu, C, Gu, C, He, X, Wang, X, Huang, S, Yuan, Q, Wu, K, Hu, W, Huang, Z, Liu, J, Wang, Z, Jia, F, Xia, K, Liu, P, Wang, X, Song, B, Zheng, J, Jiang, H, Cheng, X, Jiang, Y, Deng, S.J, Xu, H.E. | | Deposit date: | 2022-01-23 | | Release date: | 2022-03-23 | | Method: | ELECTRON MICROSCOPY (2.69 Å) | | Cite: | Structures of the Omicron spike trimer with ACE2 and an anti-Omicron antibody.
Science, 375, 2022
|
|
3EQR
 
 | | Crystal Structure of Ack1 with compound T74 | | Descriptor: | Activated CDC42 kinase 1, CHLORIDE ION, N~3~-(2,6-dimethylphenyl)-1-(3-methoxy-3-methylbutyl)-N~6~-(4-piperazin-1-ylphenyl)-1H-pyrazolo[3,4-d]pyrimidine-3,6-diamine | | Authors: | Liu, J, Wang, Z, Walker, N.P.C. | | Deposit date: | 2008-10-01 | | Release date: | 2008-12-02 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Identification and optimization of N3,N6-diaryl-1H-pyrazolo[3,4-d]pyrimidine-3,6-diamines as a novel class of ACK1 inhibitors.
Bioorg.Med.Chem.Lett., 18, 2008
|
|
2KGI
 
 | |
4LRV
 
 | | Crystal structure of DndE from Escherichia coli B7A involved in DNA phosphorothioation modification | | Descriptor: | DNA sulfur modification protein DndE | | Authors: | Hu, W, Wang, C.K, Liang, J.D, Zhang, T.L, Yang, M, Hu, Z.P, Wang, Z.J, Lan, W.X, Wu, H.M, Ding, J.P, Wu, G, Deng, Z.X, Cao, C. | | Deposit date: | 2013-07-21 | | Release date: | 2013-08-28 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural insights into DndE from Escherichia coli B7A involved in DNA phosphorothioation modification
Cell Res., 22, 2012
|
|
9EWX
 
 | | Cryo-EM structure of the Pseudomonas aeruginosa PAO1 Type IV pilus | | Descriptor: | Pilin | | Authors: | Ochner, H, Boehning, J, Wang, Z, Tarafder, A, Caspy, I, Bharat, T.A.M. | | Deposit date: | 2024-04-05 | | Release date: | 2024-05-01 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.17 Å) | | Cite: | Structure of the Pseudomonas aeruginosa PAO1 Type-IV pilus
To Be Published
|
|
3P4Z
 
 | | Time-dependent and Protein-directed In Situ Growth of Gold Nanoparticles in a Single Crystal of Lysozyme | | Descriptor: | CHLORIDE ION, GOLD 3+ ION, GOLD ION, ... | | Authors: | Wei, H, Wang, Z, Zhang, J, House, S, Gao, Y.-G, Yang, L, Robinson, H, Tan, L.H, Xing, H, Hou, C, Robertson, I.M, Zuo, J.-M, Lu, Y. | | Deposit date: | 2010-10-07 | | Release date: | 2011-02-09 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Time-dependent, protein-directed growth of gold nanoparticles within a single crystal of lysozyme.
Nat Nanotechnol, 6, 2011
|
|
8GYX
 
 | | Cryo-EM structure of human CEPT1 | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, Choline/ethanolaminephosphotransferase 1, MAGNESIUM ION | | Authors: | Qian, H.W, Wang, Z.H. | | Deposit date: | 2022-09-24 | | Release date: | 2023-03-22 | | Last modified: | 2023-10-04 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structural basis for catalysis of human choline/ethanolamine phosphotransferase 1.
Nat Commun, 14, 2023
|
|
3P64
 
 | | Time-dependent and Protein-directed In Situ Growth of Gold Nanoparticles in a Single Crystal of Lysozyme | | Descriptor: | CHLORIDE ION, GOLD 3+ ION, GOLD ION, ... | | Authors: | Wei, H, Wang, Z, Zhang, J, House, S, Gao, Y.-G, Yang, L, Robinson, H, Tan, L.H, Xing, H, Hou, C, Robertson, I.M, Zuo, J.-M, Lu, Y. | | Deposit date: | 2010-10-11 | | Release date: | 2011-02-09 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Time-dependent, protein-directed growth of gold nanoparticles within a single crystal of lysozyme.
Nat Nanotechnol, 6, 2011
|
|
8GYW
 
 | | Cryo-EM structure of human CEPT1 complexed with CDP-choline | | Descriptor: | Choline/ethanolaminephosphotransferase 1, MAGNESIUM ION, [2-CYTIDYLATE-O'-PHOSPHONYLOXYL]-ETHYL-TRIMETHYL-AMMONIUM | | Authors: | Qian, H.W, Wang, Z.H. | | Deposit date: | 2022-09-24 | | Release date: | 2023-03-22 | | Last modified: | 2023-10-04 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structural basis for catalysis of human choline/ethanolamine phosphotransferase 1.
Nat Commun, 14, 2023
|
|
3PY3
 
 | |
8HW5
 
 | |
3O72
 
 | | Crystal structure of EfeB in complex with heme | | Descriptor: | OXYGEN MOLECULE, PROTOPORPHYRIN IX CONTAINING FE, Redox component of a tripartite ferrous iron transporter | | Authors: | Liu, X, Du, Q, Wang, Z, Zhu, D, Huang, Y, Li, N, Xu, S, Gu, L. | | Deposit date: | 2010-07-30 | | Release date: | 2011-03-16 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structure and biochemical features of EfeB/YcdB from Escherichia coli O157: ASP235 plays divergent roles in different enzyme-catalyzed processes
J.Biol.Chem., 286, 2011
|
|
