7T0B
 
 | | Cryptococcus neoformans protein farnesyltransferase in complex with FPP and inhibitor 2g | | Descriptor: | (2R)-3-(cyclohexylamino)-2-hydroxypropane-1-sulfonic acid, (5S)-5-butyl-4-({1-[(4-fluorophenyl)methyl]-1H-imidazol-5-yl}methyl)-1-[3-(trifluoromethoxy)phenyl]piperazin-2-one, 1,2-ETHANEDIOL, ... | | Authors: | Wang, Y, Shi, Y, Beese, L.S. | | Deposit date: | 2021-11-29 | | Release date: | 2022-11-09 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.026 Å) | | Cite: | Structure-Guided Discovery of Potent Antifungals that Prevent Ras Signaling by Inhibiting Protein Farnesyltransferase.
J.Med.Chem., 65, 2022
|
|
7T0E
 
 | | Cryptococcus neoformans protein farnesyltransferase in complex with FPP and inhibitor 2b | | Descriptor: | (2R)-3-(cyclohexylamino)-2-hydroxypropane-1-sulfonic acid, (5S)-4-({1-[(4-bromophenyl)methyl]-1H-imidazol-5-yl}methyl)-5-butyl-1-(3-chlorophenyl)piperazin-2-one, 1,2-ETHANEDIOL, ... | | Authors: | Wang, Y, Shi, Y, Beese, L.S. | | Deposit date: | 2021-11-29 | | Release date: | 2022-11-09 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.223 Å) | | Cite: | Structure-Guided Discovery of Potent Antifungals that Prevent Ras Signaling by Inhibiting Protein Farnesyltransferase.
J.Med.Chem., 65, 2022
|
|
7T0A
 
 | | Cryptococcus neoformans protein farnesyltransferase co-crystallized with FPP and inhibitor 2f | | Descriptor: | (2R)-3-(cyclohexylamino)-2-hydroxypropane-1-sulfonic acid, (5S)-4-({1-[(4-bromophenyl)methyl]-1H-imidazol-5-yl}methyl)-5-butyl-1-[3-(trifluoromethoxy)phenyl]piperazin-2-one, 1,2-ETHANEDIOL, ... | | Authors: | Wang, Y, Shi, Y, Beese, L.S. | | Deposit date: | 2021-11-29 | | Release date: | 2022-11-09 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.099 Å) | | Cite: | Structure-Guided Discovery of Potent Antifungals that Prevent Ras Signaling by Inhibiting Protein Farnesyltransferase.
J.Med.Chem., 65, 2022
|
|
7T09
 
 | | Cryptococcus neoformans protein farnesyltransferase in complex with FPP and inhibitor 2d | | Descriptor: | (2R)-3-(cyclohexylamino)-2-hydroxypropane-1-sulfonic acid, 1,2-ETHANEDIOL, 4-{[5-({(2S)-2-butyl-5-oxo-4-[3-(trifluoromethoxy)phenyl]piperazin-1-yl}methyl)-1H-imidazol-1-yl]methyl}benzonitrile, ... | | Authors: | Wang, Y, Shi, Y, Beese, L.S. | | Deposit date: | 2021-11-29 | | Release date: | 2022-11-09 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.984 Å) | | Cite: | Structure-Guided Discovery of Potent Antifungals that Prevent Ras Signaling by Inhibiting Protein Farnesyltransferase.
J.Med.Chem., 65, 2022
|
|
7T08
 
 | | Cryptococcus neoformans protein farnesyltransferase in complex with FPP and inhibitor 2q | | Descriptor: | (2R)-3-(cyclohexylamino)-2-hydroxypropane-1-sulfonic acid, 1,2-ETHANEDIOL, 4-{[5-({(2S)-2-butyl-5-oxo-4-[3-(trifluoromethoxy)phenyl]piperazin-1-yl}methyl)-1H-imidazol-1-yl]methyl}-2-fluorobenzonitrile, ... | | Authors: | Wang, Y, Shi, Y, Beese, L.S. | | Deposit date: | 2021-11-29 | | Release date: | 2022-11-09 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.803 Å) | | Cite: | Structure-Guided Discovery of Potent Antifungals that Prevent Ras Signaling by Inhibiting Protein Farnesyltransferase.
J.Med.Chem., 65, 2022
|
|
2F90
 
 | | Crystal structure of bisphosphoglycerate mutase in complex with 3-phosphoglycerate and AlF4- | | Descriptor: | 3-PHOSPHOGLYCERIC ACID, Bisphosphoglycerate mutase, TETRAFLUOROALUMINATE ION | | Authors: | Wang, Y, Liu, L, Wei, Z, Gong, W. | | Deposit date: | 2005-12-05 | | Release date: | 2006-10-24 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Seeing the process of histidine phosphorylation in human bisphosphoglycerate mutase
J.Biol.Chem., 281, 2006
|
|
8Y1L
 
 | |
7N19
 
 | |
6VI8
 
 | | Observing a ring-cleaving dioxygenase in action through a crystalline lens - a superoxo bound structure | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 3-HYDROXYANTHRANILIC ACID, 3-hydroxyanthranilate 3,4-dioxygenase, ... | | Authors: | Wang, Y, Liu, F, Yang, Y, Liu, A. | | Deposit date: | 2020-01-12 | | Release date: | 2020-07-29 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Observing 3-hydroxyanthranilate-3,4-dioxygenase in action through a crystalline lens.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6VIA
 
 | | Observing a ring-cleaving dioxygenase in action through a crystalline lens - a seven-membered lactone bound structure | | Descriptor: | (2R,3E)-2-hydroxy-3-imino-2,3-dihydrooxepine-4-carboxylic acid, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 3-hydroxyanthranilate 3,4-dioxygenase, ... | | Authors: | Wang, Y, Liu, F, Yang, Y, Liu, A. | | Deposit date: | 2020-01-12 | | Release date: | 2020-07-29 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.591 Å) | | Cite: | Observing 3-hydroxyanthranilate-3,4-dioxygenase in action through a crystalline lens.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6VI6
 
 | | Observing a ring-cleaving dioxygenase in action through a crystalline lens - a substrate monodentately bound structure | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 3-HYDROXYANTHRANILIC ACID, 3-hydroxyanthranilate 3,4-dioxygenase, ... | | Authors: | Wang, Y, Liu, F, Yang, Y, Liu, A. | | Deposit date: | 2020-01-12 | | Release date: | 2020-07-29 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.901 Å) | | Cite: | Observing 3-hydroxyanthranilate-3,4-dioxygenase in action through a crystalline lens.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6VI7
 
 | | Probing extradiol dioxygenase mechanism in NAD(+) biosynthesis by viewing reaction cycle intermediates - a substrate bidentately bound structure | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 3-HYDROXYANTHRANILIC ACID, 3-hydroxyanthranilate 3,4-dioxygenase, ... | | Authors: | Wang, Y, Liu, F, Yang, Y, Liu, A. | | Deposit date: | 2020-01-12 | | Release date: | 2020-02-12 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.617 Å) | | Cite: | Observing 3-hydroxyanthranilate-3,4-dioxygenase in action through a crystalline lens.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6VIB
 
 | | Observing a ring-cleaving dioxygenase in action through a crystalline lens - enol tautomers of ACMS bidentately bound structure | | Descriptor: | (2Z,3Z)-2-[(2Z)-3-hydroxyprop-2-en-1-ylidene]-3-iminobutanedioic acid, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 3-hydroxyanthranilate 3,4-dioxygenase, ... | | Authors: | Wang, Y, Liu, F, Yang, Y, Liu, A. | | Deposit date: | 2020-01-12 | | Release date: | 2020-07-29 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Observing 3-hydroxyanthranilate-3,4-dioxygenase in action through a crystalline lens.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6VI9
 
 | | Observing a ring-cleaving dioxygenase in action through a crystalline lens - an alkylperoxo bound structure | | Descriptor: | (5R,6Z)-5-(hydroperoxy-kappaO)-5-(hydroxy-kappaO)-6-iminocyclohexa-1,3-diene-1-carboxylato(2-)iron, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 3-hydroxyanthranilate 3,4-dioxygenase, ... | | Authors: | Wang, Y, Liu, F, Yang, Y, Liu, A. | | Deposit date: | 2020-01-12 | | Release date: | 2020-07-29 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Observing 3-hydroxyanthranilate-3,4-dioxygenase in action through a crystalline lens.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6O58
 
 | | Human MCU-EMRE complex, dimer of channel | | Descriptor: | CALCIUM ION, Calcium uniporter protein, mitochondrial, ... | | Authors: | Wang, Y, Bai, X, Jiang, Y. | | Deposit date: | 2019-03-01 | | Release date: | 2019-05-22 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structural Mechanism of EMRE-Dependent Gating of the Human Mitochondrial Calcium Uniporter.
Cell, 177, 2019
|
|
6VI5
 
 | | Observing a ring-cleaving dioxygenase in action through a crystalline lens - a resting state structure | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 3-hydroxyanthranilate 3,4-dioxygenase, CHLORIDE ION, ... | | Authors: | Wang, Y, Liu, F, Yang, Y, Liu, A. | | Deposit date: | 2020-01-12 | | Release date: | 2020-07-29 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.604 Å) | | Cite: | Observing 3-hydroxyanthranilate-3,4-dioxygenase in action through a crystalline lens.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6X11
 
 | | Observing a ring-cleaving dioxygenase in action through a crystalline lens - an enol tautomer of ACMS monodentately bound structure | | Descriptor: | (2Z,3Z)-2-[(2Z)-3-hydroxyprop-2-en-1-ylidene]-3-iminobutanedioic acid, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 3-hydroxyanthranilate 3,4-dioxygenase, ... | | Authors: | Wang, Y, Liu, F, Yang, Y, Liu, A. | | Deposit date: | 2020-05-17 | | Release date: | 2020-07-29 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.097 Å) | | Cite: | Observing 3-hydroxyanthranilate-3,4-dioxygenase in action through a crystalline lens.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
1M2S
 
 | | Solution Structure of A New Potassium Channels Blocker from the Venom of Chinese Scorpion Buthus martensi Karsch | | Descriptor: | Toxin BmTX3 | | Authors: | Wang, Y, Li, M, Zhang, N, Wu, G, Hu, G, Wu, H. | | Deposit date: | 2002-06-25 | | Release date: | 2004-04-06 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | The solution structure of BmTx3B, a member of the scorpion toxin subfamily alpha-KTx 16
Proteins, 58, 2005
|
|
4XT9
 
 | | RORgamma (263-509) complexed with GSK2435341A and SRC2 | | Descriptor: | LYS-ILE-LEU-HIS-ARG-LEU-LEU-GLN, N-[4-(2,5-dichlorophenyl)-5-phenyl-1,3-thiazol-2-yl]-2-[4-(ethylsulfonyl)phenyl]acetamide, Nuclear receptor ROR-gamma, ... | | Authors: | Wang, Y, Ma, Y. | | Deposit date: | 2015-01-23 | | Release date: | 2015-08-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Discovery of N-(4-aryl-5-aryloxy-thiazol-2-yl)-amides as potent ROR gamma t inverse agonists
Bioorg.Med.Chem., 23, 2015
|
|
6LYH
 
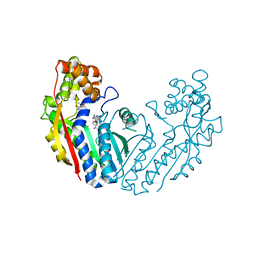 | | Crystal structure of tea N9-methyltransferase CkTcS in complex with SAH and 1,3,7-trimethyluric acid | | Descriptor: | 1,3,7-trimethyl-9H-purine-2,6,8-trione, N-methyltransferase CkTcS, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Wang, Y, Zhang, Z.-M. | | Deposit date: | 2020-02-14 | | Release date: | 2020-03-04 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.15000319 Å) | | Cite: | Identification and characterization of N9-methyltransferase involved in converting caffeine into non-stimulatory theacrine in tea.
Nat Commun, 11, 2020
|
|
5C8Y
 
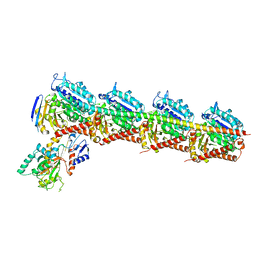 | | Crystal structure of T2R-TTL-Plinabulin complex | | Descriptor: | (3Z,6Z)-3-benzylidene-6-[(5-tert-butyl-1H-imidazol-4-yl)methylidene]piperazine-2,5-dione, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, ... | | Authors: | Wang, Y, Yu, Y, Chen, Q, Yang, J. | | Deposit date: | 2015-06-26 | | Release date: | 2015-11-04 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.594 Å) | | Cite: | Structures of a diverse set of colchicine binding site inhibitors in complex with tubulin provide a rationale for drug discovery.
Febs J., 283, 2016
|
|
5CA1
 
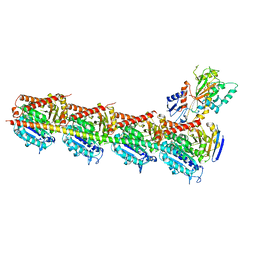 | | Crystal structure of T2R-TTL-Nocodazole complex | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, GLYCEROL, ... | | Authors: | Wang, Y, Yu, Y, Chen, Q, Yang, J. | | Deposit date: | 2015-06-29 | | Release date: | 2015-11-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.401 Å) | | Cite: | Structures of a diverse set of colchicine binding site inhibitors in complex with tubulin provide a rationale for drug discovery.
Febs J., 283, 2016
|
|
6LYI
 
 | |
2R8I
 
 | | Selectivity of Nucleoside Triphosphate Incorporation Opposite 1,N2-Propanodeoxyguanosine (PdG) by the Sulfolobus solfataricus DNA Polymerase Dpo4 Polymerase | | Descriptor: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, CALCIUM ION, DNA (5'-D(*DGP*DGP*DGP*DGP*DGP*DAP*DAP*DGP*DGP*DAP*DTP*DTP*DC)-3'), ... | | Authors: | Wang, Y, Saleh, S, Marnette, L.J, Egli, M, Stone, M.P. | | Deposit date: | 2007-09-10 | | Release date: | 2008-07-22 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Insertion of dNTPs opposite the 1,N2-propanodeoxyguanosine adduct by Sulfolobus solfataricus P2 DNA polymerase IV
Biochemistry, 47, 2008
|
|
5CA0
 
 | | Crystal structure of T2R-TTL-Lexibulin complex | | Descriptor: | 1-ethyl-3-[2-methoxy-4-(5-methyl-4-{[(1S)-1-(pyridin-3-yl)butyl]amino}pyrimidin-2-yl)phenyl]urea, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, ... | | Authors: | Wang, Y, Yu, Y, Chen, Q, Yang, J. | | Deposit date: | 2015-06-29 | | Release date: | 2015-11-04 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.501 Å) | | Cite: | Structures of a diverse set of colchicine binding site inhibitors in complex with tubulin provide a rationale for drug discovery.
Febs J., 283, 2016
|
|
