8JD6
 
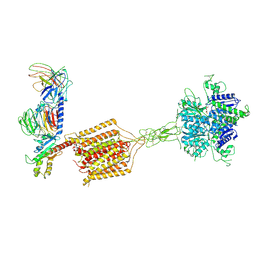 | | Cryo-EM structure of Gi1-bound metabotropic glutamate receptor mGlu4 | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(i) subunit alpha-3, ... | | Authors: | Wang, X, Wang, M, Xu, T, Feng, Y, Han, S, Lin, S, Zhao, Q, Wu, B. | | Deposit date: | 2023-05-12 | | Release date: | 2023-06-21 | | Last modified: | 2023-10-18 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural insights into dimerization and activation of the mGlu2-mGlu3 and mGlu2-mGlu4 heterodimers.
Cell Res., 33, 2023
|
|
8JCV
 
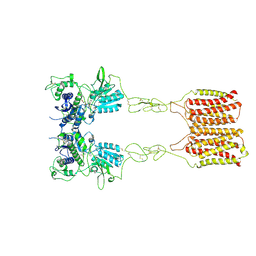 | | Cryo-EM structure of mGlu2-mGlu3 heterodimer in presence of LY341495 (dimerization mode II) | | Descriptor: | 2-[(1S,2S)-2-carboxycyclopropyl]-3-(9H-xanthen-9-yl)-D-alanine, 2-acetamido-2-deoxy-beta-D-glucopyranose, Metabotropic glutamate receptor 2,Peptidyl-prolyl cis-trans isomerase FKBP1A, ... | | Authors: | Wang, X, Wang, M, Xu, T, Feng, Y, Han, S, Zhao, Q, Wu, B. | | Deposit date: | 2023-05-12 | | Release date: | 2023-06-21 | | Last modified: | 2023-10-18 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural insights into dimerization and activation of the mGlu2-mGlu3 and mGlu2-mGlu4 heterodimers.
Cell Res., 33, 2023
|
|
8JD2
 
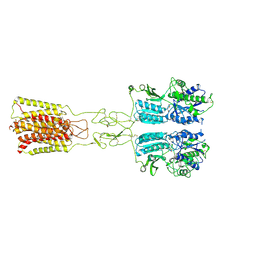 | | Cryo-EM structure of G protein-free mGlu2-mGlu3 heterodimer in Acc state | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GLUTAMIC ACID, Metabotropic glutamate receptor 2,Peptidyl-prolyl cis-trans isomerase FKBP1A, ... | | Authors: | Wang, X, Wang, M, Xu, T, Feng, Y, Han, S, Lin, S, Zhao, Q, Wu, B. | | Deposit date: | 2023-05-12 | | Release date: | 2023-06-21 | | Last modified: | 2023-10-18 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structural insights into dimerization and activation of the mGlu2-mGlu3 and mGlu2-mGlu4 heterodimers.
Cell Res., 33, 2023
|
|
8JD5
 
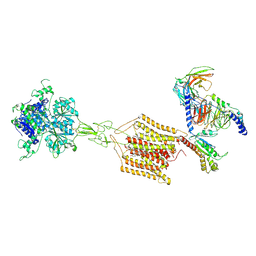 | | Cryo-EM structure of Gi1-bound mGlu2-mGlu4 heterodimer | | Descriptor: | 1-butyl-3-chloranyl-4-(4-phenylpiperidin-1-yl)pyridin-2-one, 2-acetamido-2-deoxy-beta-D-glucopyranose, 5-methyl-N-(4-methylpyrimidin-2-yl)-4-(1H-pyrazol-4-yl)-1,3-thiazol-2-amine, ... | | Authors: | Wang, X, Wang, M, Xu, T, Feng, Y, Han, S, Zhao, Q, Wu, B. | | Deposit date: | 2023-05-12 | | Release date: | 2023-06-21 | | Last modified: | 2023-10-18 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural insights into dimerization and activation of the mGlu2-mGlu3 and mGlu2-mGlu4 heterodimers.
Cell Res., 33, 2023
|
|
8JCU
 
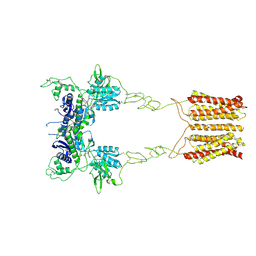 | | Cryo-EM structure of mGlu2-mGlu3 heterodimer in presence of LY341495 (dimerization mode I) | | Descriptor: | 2-[(1S,2S)-2-carboxycyclopropyl]-3-(9H-xanthen-9-yl)-D-alanine, 2-acetamido-2-deoxy-beta-D-glucopyranose, Metabotropic glutamate receptor 2,Peptidyl-prolyl cis-trans isomerase FKBP1A, ... | | Authors: | Wang, X, Wang, M, Xu, T, Feng, Y, Zhao, Q, Wu, B. | | Deposit date: | 2023-05-12 | | Release date: | 2023-06-21 | | Last modified: | 2023-10-18 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structural insights into dimerization and activation of the mGlu2-mGlu3 and mGlu2-mGlu4 heterodimers.
Cell Res., 33, 2023
|
|
8JD3
 
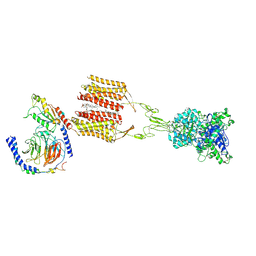 | | Cryo-EM structure of Gi1-bound mGlu2-mGlu3 heterodimer | | Descriptor: | 1-butyl-3-chloranyl-4-(4-phenylpiperidin-1-yl)pyridin-2-one, CHOLESTEROL, DI-PALMITOYL-3-SN-PHOSPHATIDYLETHANOLAMINE, ... | | Authors: | Wang, X, Wang, M, Xu, T, Feng, Y, Han, S, Lin, S, Zhao, Q, Wu, B. | | Deposit date: | 2023-05-12 | | Release date: | 2023-06-21 | | Last modified: | 2023-10-18 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural insights into dimerization and activation of the mGlu2-mGlu3 and mGlu2-mGlu4 heterodimers.
Cell Res., 33, 2023
|
|
8JD1
 
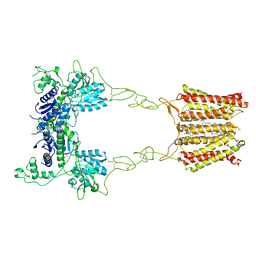 | | Cryo-EM structure of mGlu2-mGlu3 heterodimer in Rco state | | Descriptor: | CHOLESTEROL, GLUTAMIC ACID, Metabotropic glutamate receptor 2,Peptidyl-prolyl cis-trans isomerase FKBP1A, ... | | Authors: | Wang, X, Wang, M, Xu, T, Feng, Y, Han, S, Lin, S, Zhao, Q, Wu, B. | | Deposit date: | 2023-05-12 | | Release date: | 2023-06-21 | | Last modified: | 2023-10-18 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structural insights into dimerization and activation of the mGlu2-mGlu3 and mGlu2-mGlu4 heterodimers.
Cell Res., 33, 2023
|
|
8JCW
 
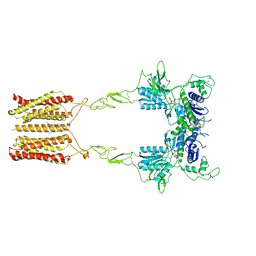 | | Cryo-EM structure of mGlu2-mGlu3 heterodimer in presence of LY341495 and NAM563 (dimerization mode I) | | Descriptor: | 2-[(1S,2S)-2-carboxycyclopropyl]-3-(9H-xanthen-9-yl)-D-alanine, 2-acetamido-2-deoxy-beta-D-glucopyranose, CHOLESTEROL, ... | | Authors: | Wang, X, Wang, M, Xu, T, Feng, Y, Han, S, Lin, S, Zhao, Q, Wu, B. | | Deposit date: | 2023-05-12 | | Release date: | 2023-06-21 | | Last modified: | 2023-10-18 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural insights into dimerization and activation of the mGlu2-mGlu3 and mGlu2-mGlu4 heterodimers.
Cell Res., 33, 2023
|
|
8JD4
 
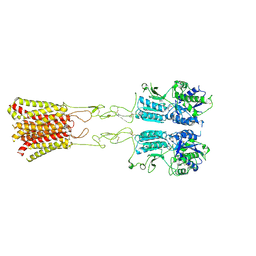 | | Cryo-EM structure of G protein-free mGlu2-mGlu4 heterodimer in Acc state | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GLUTAMIC ACID, Metabotropic glutamate receptor 2,Peptidyl-prolyl cis-trans isomerase FKBP1A, ... | | Authors: | Wang, X, Wang, M, Xu, T, Feng, Y, Han, S, Lin, S, Zhao, Q, Wu, B. | | Deposit date: | 2023-05-12 | | Release date: | 2023-06-21 | | Last modified: | 2023-10-18 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural insights into dimerization and activation of the mGlu2-mGlu3 and mGlu2-mGlu4 heterodimers.
Cell Res., 33, 2023
|
|
8JCY
 
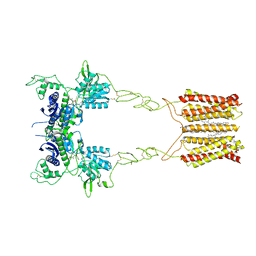 | | Cryo-EM structure of mGlu2-mGlu3 heterodimer in presence of LY341495, NAM563, and LY2389575 (dimerization mode I) | | Descriptor: | 2-[(1S,2S)-2-carboxycyclopropyl]-3-(9H-xanthen-9-yl)-D-alanine, 2-acetamido-2-deoxy-beta-D-glucopyranose, CHOLESTEROL, ... | | Authors: | Wang, X, Wang, M, Xu, T, Feng, Y, Han, S, Lin, S, Zhao, Q, Wu, B. | | Deposit date: | 2023-05-12 | | Release date: | 2023-06-21 | | Last modified: | 2023-10-18 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural insights into dimerization and activation of the mGlu2-mGlu3 and mGlu2-mGlu4 heterodimers.
Cell Res., 33, 2023
|
|
8JCZ
 
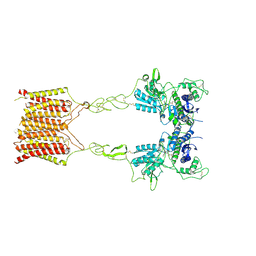 | | Cryo-EM structure of mGlu2-mGlu3 heterodimer in presence of LY341495, NAM563, and LY2389575 (dimerization mode III) | | Descriptor: | 2-[(1S,2S)-2-carboxycyclopropyl]-3-(9H-xanthen-9-yl)-D-alanine, 2-acetamido-2-deoxy-beta-D-glucopyranose, Metabotropic glutamate receptor 2,Peptidyl-prolyl cis-trans isomerase FKBP1A, ... | | Authors: | Wang, X, Wang, M, Xu, T, Feng, Y, Han, S, Lin, S, Zhao, Q, Wu, B. | | Deposit date: | 2023-05-12 | | Release date: | 2023-06-21 | | Last modified: | 2023-10-18 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural insights into dimerization and activation of the mGlu2-mGlu3 and mGlu2-mGlu4 heterodimers.
Cell Res., 33, 2023
|
|
8JCX
 
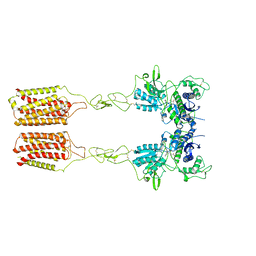 | | Cryo-EM structure of mGlu2-mGlu3 heterodimer in presence of LY341495 and NAM563 (dimerization mode II) | | Descriptor: | 2-[(1S,2S)-2-carboxycyclopropyl]-3-(9H-xanthen-9-yl)-D-alanine, 2-acetamido-2-deoxy-beta-D-glucopyranose, Metabotropic glutamate receptor 2,Peptidyl-prolyl cis-trans isomerase FKBP1A, ... | | Authors: | Wang, X, Wang, M, Xu, T, Feng, Y, Han, S, Lin, S, Zhao, Q, Wu, B. | | Deposit date: | 2023-05-12 | | Release date: | 2023-06-21 | | Last modified: | 2023-10-18 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural insights into dimerization and activation of the mGlu2-mGlu3 and mGlu2-mGlu4 heterodimers.
Cell Res., 33, 2023
|
|
8JD0
 
 | | Cryo-EM structure of mGlu2-mGlu3 heterodimer in presence of NAM563 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(1-methylpyrazol-4-yl)-7-[[(2~{S})-2-(trifluoromethyl)morpholin-4-yl]methyl]quinoline-2-carboxamide, CHOLESTEROL, ... | | Authors: | Wang, X, Wang, M, Xu, T, Feng, Y, Han, S, Lin, S, Zhao, Q, Wu, B. | | Deposit date: | 2023-05-12 | | Release date: | 2023-06-21 | | Last modified: | 2023-10-18 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural insights into dimerization and activation of the mGlu2-mGlu3 and mGlu2-mGlu4 heterodimers.
Cell Res., 33, 2023
|
|
5X6O
 
 | | Intact ATR/Mec1-ATRIP/Ddc2 complex | | Descriptor: | DNA damage checkpoint protein LCD1, Serine/threonine-protein kinase MEC1 | | Authors: | Wang, X, Ran, T, Cai, G. | | Deposit date: | 2017-02-22 | | Release date: | 2017-12-20 | | Last modified: | 2019-11-06 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | 3.9 angstrom structure of the yeast Mec1-Ddc2 complex, a homolog of human ATR-ATRIP.
Science, 358, 2017
|
|
2L9H
 
 | | Oligomeric Structure of the Chemokine CCL5/RANTES from NMR, MS, and SAXS Data | | Descriptor: | C-C motif chemokine 5 | | Authors: | Wang, X, Watson, C.M, Sharp, J.S, Handel, T.M, Prestegard, J.H. | | Deposit date: | 2011-02-09 | | Release date: | 2011-06-22 | | Last modified: | 2011-08-24 | | Method: | SOLUTION NMR, SOLUTION SCATTERING | | Cite: | Oligomeric Structure of the Chemokine CCL5/RANTES from NMR, MS, and SAXS Data.
Structure, 19, 2011
|
|
6AJ8
 
 | | Crystal structure of Trypanosoma brucei glycosomal isocitrate dehydrogenase in complex with NADP+, alpha-ketoglutarate and ca2+ | | Descriptor: | 2-OXOGLUTARIC ACID, CALCIUM ION, Isocitrate dehydrogenase [NADP], ... | | Authors: | Wang, X, Inaoka, D.K, Shiba, T, Balogun, E.O, Ziebart, N, Allman, S, Watanabe, Y, Nozaki, T, Boshart, M, Bringaud, F, Harada, S, Kita, K. | | Deposit date: | 2018-08-27 | | Release date: | 2019-08-28 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Biochemical characterization of a novel Trypanosoma brucei glycosomal isocitrate dehydrogenase with dual coenzyme specificity (NADP+/NAD+)
To Be Published
|
|
5DQR
 
 | |
5ZUD
 
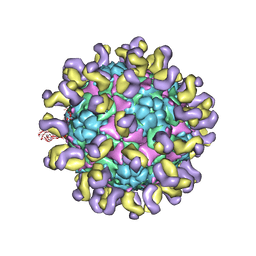 | | Fit R10 Fab coordinates into the cryo-EM of EV71 in complex with D6 | | Descriptor: | Capsid protein VP1, R10 ANTIBODY HEAVY CHAIN, R10 ANTIBODY LIGHT CHAIN, ... | | Authors: | Wang, X, Zhu, L, Wang, N. | | Deposit date: | 2018-05-07 | | Release date: | 2019-12-25 | | Last modified: | 2021-01-06 | | Method: | ELECTRON MICROSCOPY (4.9 Å) | | Cite: | Neutralization Mechanisms of Two Highly Potent Antibodies against Human Enterovirus 71.
Mbio, 9, 2018
|
|
5ZUF
 
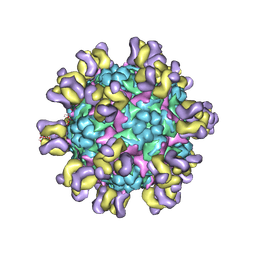 | | Fit R10 Fab coordinates into the cryo-EM of EV71 in complex with A9 | | Descriptor: | Capsid protein VP1, R10 ANTIBODY HEAVY CHAIN, R10 ANTIBODY LIGHT CHAIN, ... | | Authors: | Wang, X, Zhu, L, Wang, N. | | Deposit date: | 2018-05-07 | | Release date: | 2019-12-25 | | Last modified: | 2021-01-06 | | Method: | ELECTRON MICROSCOPY (6.8 Å) | | Cite: | Neutralization Mechanisms of Two Highly Potent Antibodies against Human Enterovirus 71.
Mbio, 9, 2018
|
|
6M16
 
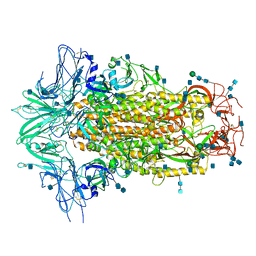 | | Cryo-EM structures of SADS-CoV spike glycoproteins | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Wang, X, Yu, J, Qiao, S, Guo, R. | | Deposit date: | 2020-02-24 | | Release date: | 2020-05-27 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (2.83 Å) | | Cite: | Cryo-EM structures of HKU2 and SADS-CoV spike glycoproteins provide insights into coronavirus evolution.
Nat Commun, 11, 2020
|
|
6M15
 
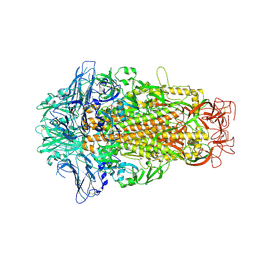 | | Cryo-EM structures of HKU2 spike glycoproteins | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Wang, X, Yu, J, Qiao, S, Guo, R. | | Deposit date: | 2020-02-24 | | Release date: | 2020-05-27 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (2.38 Å) | | Cite: | Cryo-EM structures of HKU2 and SADS-CoV spike glycoproteins provide insights into coronavirus evolution.
Nat Commun, 11, 2020
|
|
5HLZ
 
 | |
6NUC
 
 | | Structure of Calcineurin in complex with NHE1 peptide | | Descriptor: | CALCIUM ION, Calcineurin subunit B type 1, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Wang, X, Page, R, Peti, W. | | Deposit date: | 2019-01-31 | | Release date: | 2019-07-03 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Molecular basis for the binding and selective dephosphorylation of Na+/H+exchanger 1 by calcineurin.
Nat Commun, 10, 2019
|
|
5HLY
 
 | |
6NUU
 
 | | Structure of Calcineurin mutant in complex with NHE1 peptide | | Descriptor: | CALCIUM ION, Calcineurin subunit B type 1, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Wang, X, Page, R, Peti, W. | | Deposit date: | 2019-02-02 | | Release date: | 2019-07-03 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Molecular basis for the binding and selective dephosphorylation of Na+/H+exchanger 1 by calcineurin.
Nat Commun, 10, 2019
|
|
