7EB2
 
 | | Cryo-EM structure of human GABA(B) receptor-Gi protein complex | | Descriptor: | (3S)-5,7-ditert-butyl-3-oxidanyl-3-(trifluoromethyl)-1-benzofuran-2-one, Gamma-aminobutyric acid type B receptor subunit 1, Gamma-aminobutyric acid type B receptor subunit 2, ... | | Authors: | Shen, C, Mao, C, Xu, C, Jin, N, Zhang, H, Shen, D, Shen, Q, Wang, X, Hou, T, Rondard, P, Chen, Z, Pin, J, Zhang, Y, Liu, J. | | Deposit date: | 2021-03-08 | | Release date: | 2021-05-05 | | Last modified: | 2021-07-07 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural basis of GABA B receptor-G i protein coupling.
Nature, 594, 2021
|
|
2O2N
 
 | | Solution structure of the anti-apoptotic protein Bcl-xL in complex with an acyl-sulfonamide-based ligand | | Descriptor: | 4-[4-(BIPHENYL-2-YLMETHYL)PIPERAZIN-1-YL]-N-[(4-{[1,1-DIMETHYL-2-(PHENYLTHIO)ETHYL]AMINO}-3-NITROPHENYL)SULFONYL]BENZAMIDE, Apoptosis regulator Bcl-X | | Authors: | Bruncko, M, Oost, T.K, Belli, B.A, Ding, H, Joseph, M.K, Kunzer, A, Martineau, D, McClellan, W.J, Mitten, M, Ng, S.C, Nimmer, P.M, Oltersdorf, T, Park, C.M, Petros, A.M, Shoemaker, A.R, Song, X, Wang, X, Wendt, M.D, Zhang, H, Fesik, S.W, Rosenberg, S.H, Elmore, S.W. | | Deposit date: | 2006-11-30 | | Release date: | 2007-02-27 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Studies Leading to Potent, Dual Inhibitors of Bcl-2 and Bcl-xL.
J.Med.Chem., 50, 2007
|
|
6J11
 
 | | MERS-CoV spike N-terminal domain and 7D10 scFv complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, N-terminal domain of Spike glycoprotein, ... | | Authors: | Zhou, H, Zhang, S, Zhang, S, Tang, W, Wang, X. | | Deposit date: | 2018-12-27 | | Release date: | 2019-07-24 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural definition of a neutralization epitope on the N-terminal domain of MERS-CoV spike glycoprotein.
Nat Commun, 10, 2019
|
|
2O21
 
 | | Solution structure of the anti-apoptotic protein Bcl-2 in complex with an acyl-sulfonamide-based ligand | | Descriptor: | 3-NITRO-N-{4-[2-(2-PHENYLETHYL)-1,3-BENZOTHIAZOL-5-YL]BENZOYL}-4-{[2-(PHENYLSULFANYL)ETHYL]AMINO}BENZENESULFONAMIDE, Apoptosis regulator Bcl-2 | | Authors: | Bruncko, M, Oost, T.K, Belli, B.A, Ding, H, Joseph, M.K, Kunzer, A, Martineau, D, McClellan, W.J, Mitten, M, Ng, S.C, Nimmer, P.M, Oltersdorf, T, Park, C.M, Petros, A.M, Shoemaker, A.R, Song, X, Wang, X, Wendt, M.D, Zhang, H, Fesik, S.W, Rosenberg, S.H, Elmore, S.W. | | Deposit date: | 2006-11-29 | | Release date: | 2007-02-27 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Studies Leading to Potent, Dual Inhibitors of Bcl-2 and Bcl-xL.
J.Med.Chem., 50, 2007
|
|
2O2F
 
 | | Solution structure of the anti-apoptotic protein Bcl-2 in complex with an acyl-sulfonamide-based ligand | | Descriptor: | 4-(4-BENZYL-4-METHOXYPIPERIDIN-1-YL)-N-[(4-{[1,1-DIMETHYL-2-(PHENYLTHIO)ETHYL]AMINO}-3-NITROPHENYL)SULFONYL]BENZAMIDE, Apoptosis regulator Bcl-2 | | Authors: | Bruncko, M, Oost, T.K, Belli, B.A, Ding, H, Joseph, M.K, Kunzer, A, Martineau, D, McClellan, W.J, Mitten, M, Ng, S.C, Nimmer, P.M, Oltersdorf, T, Park, C.M, Petros, A.M, Shoemaker, A.R, Song, X, Wang, X, Wendt, M.D, Zhang, H, Fesik, S.W, Rosenberg, S.H, Elmore, S.W. | | Deposit date: | 2006-11-29 | | Release date: | 2007-02-27 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Studies Leading to Potent, Dual Inhibitors of Bcl-2 and Bcl-xL.
J.Med.Chem., 50, 2007
|
|
7YCX
 
 | | The structure of INTAC-PEC complex | | Descriptor: | DNA-directed RNA polymerase II subunit E, DNA-directed RNA polymerase II subunit F, DNA-directed RNA polymerase II subunit RPB1,DNA-directed RNA polymerase II subunit RPB1, ... | | Authors: | Zheng, H, Jin, Q, Wang, X, Qi, Y, Liu, W, Ren, Y, Zhao, D, Chen, F.X, Cheng, J, Chen, X, Xu, Y. | | Deposit date: | 2022-07-02 | | Release date: | 2023-03-15 | | Last modified: | 2023-09-27 | | Method: | ELECTRON MICROSCOPY (4.18 Å) | | Cite: | Structural basis of INTAC-regulated transcription.
Protein Cell, 14, 2023
|
|
6LTJ
 
 | | Structure of nucleosome-bound human BAF complex | | Descriptor: | AT-rich interactive domain-containing protein 1A, Actin, cytoplasmic 1, ... | | Authors: | He, S, Wu, Z, Tian, Y, Yu, Z, Yu, J, Wang, X, Li, J, Liu, B, Xu, Y. | | Deposit date: | 2020-01-22 | | Release date: | 2020-02-12 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structure of nucleosome-bound human BAF complex.
Science, 367, 2020
|
|
2O1Y
 
 | | Solution structure of the anti-apoptotic protein Bcl-xL in complex with an acyl-sulfonamide-based ligand | | Descriptor: | 3-NITRO-N-{4-[2-(2-PHENYLETHYL)-1,3-BENZOTHIAZOL-5-YL]BENZOYL}-4-{[2-(PHENYLSULFANYL)ETHYL]AMINO}BENZENESULFONAMIDE, Apoptosis regulator Bcl-X | | Authors: | Bruncko, M, Oost, T.K, Belli, B.A, Ding, H, Joseph, M.K, Kunzer, A, Martineau, D, McClellan, W.J, Mitten, M, Ng, S.C, Nimmer, P.M, Oltersdorf, T, Park, C.M, Petros, A.M, Shoemaker, A.R, Song, X, Wang, X, Wendt, M.D, Zhang, H, Fesik, S.W, Rosenberg, S.H, Elmore, S.W. | | Deposit date: | 2006-11-29 | | Release date: | 2007-02-27 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Studies Leading to Potent, Dual Inhibitors of Bcl-2 and Bcl-xL.
J.Med.Chem., 50, 2007
|
|
2O22
 
 | | Solution structure of the anti-apoptotic protein Bcl-2 in complex with an acyl-sulfonamide-based ligand | | Descriptor: | Apoptosis regulator Bcl-2, N-[(4-{[1,1-dimethyl-2-(phenylthio)ethyl]amino}-3-nitrophenyl)sulfonyl]-4-(4,4-dimethylpiperidin-1-yl)benzamide | | Authors: | Bruncko, M, Oost, T.K, Belli, B.A, Ding, H, Joseph, M.K, Kunzer, A, Martineau, D, McClellan, W.J, Mitten, M, Ng, S.C, Nimmer, P.M, Oltersdorf, T, Park, C.M, Petros, A.M, Shoemaker, A.R, Song, X, Wang, X, Wendt, M.D, Zhang, H, Fesik, S.W, Rosenberg, S.H, Elmore, S.W. | | Deposit date: | 2006-11-29 | | Release date: | 2007-02-27 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Studies Leading to Potent, Dual Inhibitors of Bcl-2 and Bcl-xL.
J.Med.Chem., 50, 2007
|
|
2O2M
 
 | | Solution structure of the anti-apoptotic protein Bcl-xL in complex with an acyl-sulfonamide-based ligand | | Descriptor: | 4-(4-BENZYL-4-METHOXYPIPERIDIN-1-YL)-N-[(4-{[1,1-DIMETHYL-2-(PHENYLTHIO)ETHYL]AMINO}-3-NITROPHENYL)SULFONYL]BENZAMIDE, Apoptosis regulator Bcl-X | | Authors: | Bruncko, M, Oost, T.K, Belli, B.A, Ding, H, Joseph, M.K, Kunzer, A, Martineau, D, McClellan, W.J, Mitten, M, Ng, S.C, Nimmer, P.M, Oltersdorf, T, Park, C.M, Petros, A.M, Shoemaker, A.R, Song, X, Wang, X, Wendt, M.D, Zhang, H, Fesik, S.W, Rosenberg, S.H, Elmore, S.W. | | Deposit date: | 2006-11-30 | | Release date: | 2007-02-27 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Studies Leading to Potent, Dual Inhibitors of Bcl-2 and Bcl-xL.
J.Med.Chem., 50, 2007
|
|
7YKS
 
 | |
7YKR
 
 | |
6MEP
 
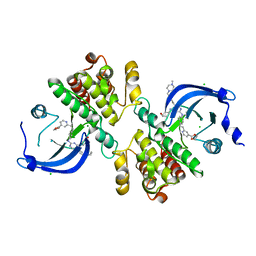 | | Crystal structure of the catalytic domain of the proto-oncogene tyrosine-protein kinase MER in complex with inhibitor UNC3437 | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, Tyrosine-protein kinase Mer, ... | | Authors: | Da, C, Zhang, D, Stashko, M.A, Cheng, A, Hunter, D, Norris-Drouin, J, Graves, L, Machius, M, Miley, M.J, DeRyckere, D, Earp, H.S, Graham, D.K, Frye, S.V, Wang, X, Kireev, D. | | Deposit date: | 2018-09-06 | | Release date: | 2019-09-11 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.893 Å) | | Cite: | Data-Driven Construction of Antitumor Agents with Controlled Polypharmacology.
J.Am.Chem.Soc., 141, 2019
|
|
2NC4
 
 | | Solution Structure of N-Galactosylated Pin1 WW Domain | | Descriptor: | Pin1 WW Domain, beta-D-galactopyranose | | Authors: | Hsu, C, Park, S, Mortenson, D.E, Foley, B, Wang, X, Woods, R.J, Case, D.A, Powers, E.T, Wong, C, Dyson, H, Kelly, J.W. | | Deposit date: | 2016-03-20 | | Release date: | 2016-06-08 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | The Dependence of Carbohydrate-Aromatic Interaction Strengths on the Structure of the Carbohydrate.
J.Am.Chem.Soc., 138, 2016
|
|
2NC5
 
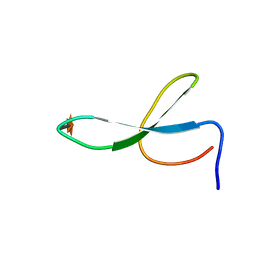 | | Solution Structure of N-Xylosylated Pin1 WW Domain | | Descriptor: | Pin1 WW Domain, beta-D-xylopyranose | | Authors: | Hsu, C, Park, S, Mortenson, D.E, Foley, B, Wang, X, Woods, R.J, Case, D.A, Powers, E.T, Wong, C, Dyson, H, Kelly, J.W. | | Deposit date: | 2016-03-20 | | Release date: | 2016-06-08 | | Last modified: | 2020-07-29 | | Method: | SOLUTION NMR | | Cite: | The Dependence of Carbohydrate-Aromatic Interaction Strengths on the Structure of the Carbohydrate.
J.Am.Chem.Soc., 138, 2016
|
|
7E3N
 
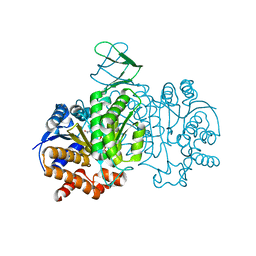 | | Crystal structure of Isocitrate dehydrogenase D252N mutant from Trypanosoma brucei in complexed with NADP+, alpha-ketoglutarate, and ca2+ | | Descriptor: | CALCIUM ION, ISOCITRIC ACID, Isocitrate dehydrogenase [NADP], ... | | Authors: | Arai, N, Shiba, T, Inaoka, D.K, Kita, K, Wang, X, Otani, M, Matsushiro, S, Kojima, C. | | Deposit date: | 2021-02-09 | | Release date: | 2022-02-16 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of Isocitrate dehydrogenase from Trypanosoma brucei.
To Be Published
|
|
2NC3
 
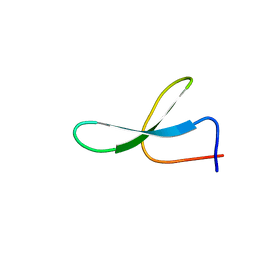 | | Solution Structure of N-Allosylated Pin1 WW Domain | | Descriptor: | Pin1 WW Domain, beta-D-allopyranose | | Authors: | Hsu, C, Park, S, Mortenson, D.E, Foley, B, Wang, X, Woods, R.J, Case, D.A, Powers, E.T, Wong, C, Dyson, H, Kelly, J.W. | | Deposit date: | 2016-03-20 | | Release date: | 2016-06-08 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The Dependence of Carbohydrate-Aromatic Interaction Strengths on the Structure of the Carbohydrate.
J.Am.Chem.Soc., 138, 2016
|
|
6KH2
 
 | |
2KS6
 
 | |
7XD9
 
 | | Crystal Structure of Dengue Virus serotype 2 (DENV2) Polymerase Elongation Complex (CTP Form) | | Descriptor: | CYTIDINE-5'-TRIPHOSPHATE, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Wu, J, Wang, X, Gong, P. | | Deposit date: | 2022-03-26 | | Release date: | 2022-12-21 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.58 Å) | | Cite: | Structural basis of transition from initiation to elongation in de novo viral RNA-dependent RNA polymerases.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
7XD8
 
 | | Crystal Structure of Dengue Virus Serotype 2 (DENV2) Polymerase Elongation Complex (Native Form) | | Descriptor: | GLYCEROL, NS5, RNA (30-mer), ... | | Authors: | Wu, J, Wang, X, Gong, P. | | Deposit date: | 2022-03-26 | | Release date: | 2022-12-21 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Structural basis of transition from initiation to elongation in de novo viral RNA-dependent RNA polymerases.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
2N6F
 
 | | Structure of Pleiotrophin | | Descriptor: | Pleiotrophin | | Authors: | Ryan, E.O, Shen, D, Wang, X. | | Deposit date: | 2015-08-20 | | Release date: | 2016-04-20 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Structural studies reveal an important role for the pleiotrophin C-terminus in mediating interactions with chondroitin sulfate.
Febs J., 283, 2016
|
|
7VYV
 
 | | Cryo-EM structure of Depo32, a Klebsiella phage depolymerase targets the K2 serotype K. pneumoniae | | Descriptor: | Depolymerase | | Authors: | Cai, R, Ren, Z, Zhao, R, Wang, X, Guo, Z, Du, R, Han, W, Ru, H, Gu, J. | | Deposit date: | 2021-11-15 | | Release date: | 2023-08-30 | | Method: | ELECTRON MICROSCOPY (2.32 Å) | | Cite: | Depo32, a depolymerase of Klebsiella phage, efficiently controls infection caused by Klebsiella pneumoniae with K2 serotype CPS
To Be Published
|
|
7VZ3
 
 | | Cryo-EM structure of Depo32, a Klebsiella phage depolymerase targets the K2 serotype K. pneumoniae | | Descriptor: | Depolymerase | | Authors: | Cai, R, Ren, Z, Zhao, R, Wang, X, Guo, Z, Du, R, Han, W, Ru, H, Gu, J. | | Deposit date: | 2021-11-15 | | Release date: | 2023-08-30 | | Method: | ELECTRON MICROSCOPY (2.46 Å) | | Cite: | Depo32, a depolymerase of Klebsiella phage, efficiently controls infection caused by Klebsiella pneumoniae with K2 serotype CPS
To Be Published
|
|
2N62
 
 | | ddFLN5+110 | | Descriptor: | gelation factor, secretion monitor chimera | | Authors: | Cabrita, L.D, Cassaignau, A.M.E, Launay, H.M.M, Waudby, C.A, Camilloni, C, Robertson, A.L, Wang, X, Wlodarski, T, Wentink, A.S, Vendruscolo, M, Dobson, C.M, Christodoulou, J. | | Deposit date: | 2015-08-10 | | Release date: | 2016-03-02 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | A structural ensemble of a ribosome-nascent chain complex during cotranslational protein folding.
Nat.Struct.Mol.Biol., 23, 2016
|
|
