4TO1
 
 | | Structure basis of cellular dNTP regulation, SAMHD1-GTP-dATP/dCTP-dCTP complex | | Descriptor: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, 2'-DEOXYCYTIDINE-5'-TRIPHOSPHATE, Deoxynucleoside triphosphate triphosphohydrolase SAMHD1, ... | | Authors: | Ji, X, Tang, C, Zhao, Q, Wang, W, Xiong, Y. | | Deposit date: | 2014-06-05 | | Release date: | 2014-10-01 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Structural basis of cellular dNTP regulation by SAMHD1.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4TNZ
 
 | | Structure basis of cellular dNTP regulation, SAMHD1-GTP-dATP-dTTP complex | | Descriptor: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, Deoxynucleoside triphosphate triphosphohydrolase SAMHD1, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Ji, X, Tang, C, Zhao, Q, Wang, W, Xiong, Y. | | Deposit date: | 2014-06-05 | | Release date: | 2014-10-01 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Structural basis of cellular dNTP regulation by SAMHD1.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4TNR
 
 | | Structure basis of cellular dNTP regulation, SAMHD1-GTP-dATP-dATP complex | | Descriptor: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, Deoxynucleoside triphosphate triphosphohydrolase SAMHD1, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Ji, X, Tang, C, Zhao, Q, Wang, W, Xiong, Y. | | Deposit date: | 2014-06-04 | | Release date: | 2014-10-01 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structural basis of cellular dNTP regulation by SAMHD1.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4TO4
 
 | | Structure basis of cellular dNTP regulation, SAMHD1-GTP-dGTP-dCTP complex | | Descriptor: | 2'-DEOXYCYTIDINE-5'-TRIPHOSPHATE, 2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, Deoxynucleoside triphosphate triphosphohydrolase SAMHD1, ... | | Authors: | Ji, X, Tang, C, Zhao, Q, Wang, W, Xiong, Y. | | Deposit date: | 2014-06-05 | | Release date: | 2014-10-01 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis of cellular dNTP regulation by SAMHD1.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4TNQ
 
 | | Structural basis of cellular dNTP regulation, SAMHD1-GTP-dTTP-dTTP complex | | Descriptor: | Deoxynucleoside triphosphate triphosphohydrolase SAMHD1, GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Ji, X, Tang, C, Zhao, Q, Wang, W, Xiong, Y. | | Deposit date: | 2014-06-04 | | Release date: | 2014-10-01 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Structural basis of cellular dNTP regulation by SAMHD1.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4TO6
 
 | | Structure basis of cellular dNTP regulation, SAMHD1-dGTP-dATP-dTTP/dGTP complex | | Descriptor: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, 2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, Deoxynucleoside triphosphate triphosphohydrolase SAMHD1, ... | | Authors: | Ji, X, Tang, C, Zhao, Q, Wang, W, Xiong, Y. | | Deposit date: | 2014-06-05 | | Release date: | 2014-10-01 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Structural basis of cellular dNTP regulation by SAMHD1.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4TNY
 
 | | Structural basis of cellular dNTP regulation, SAMHD1-dGTP-dATP-dGTP complex | | Descriptor: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, 2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, Deoxynucleoside triphosphate triphosphohydrolase SAMHD1, ... | | Authors: | Ji, X, Tang, C, Zhao, Q, Wang, W, Xiong, Y. | | Deposit date: | 2014-06-05 | | Release date: | 2014-10-01 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural basis of cellular dNTP regulation by SAMHD1.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
5D0O
 
 | | BamABCDE complex, outer membrane beta barrel assembly machinery entire complex | | Descriptor: | Outer membrane protein assembly factor BamA, Outer membrane protein assembly factor BamB, Outer membrane protein assembly factor BamC, ... | | Authors: | Gu, Y, Paterson, N, Zeng, Y, Dong, H, Wang, W, Dong, C. | | Deposit date: | 2015-08-03 | | Release date: | 2016-03-09 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural basis of outer membrane protein insertion by the BAM complex.
Nature, 531, 2016
|
|
3HVL
 
 | | Tethered PXR-LBD/SRC-1p complexed with SR-12813 | | Descriptor: | Pregnane X receptor, Linker, Steroid receptor coactivator 1, ... | | Authors: | Lesburg, C.A, Wang, W, Prosise, W.W, Chen, J, Taremi, S.S, Le, H.V, Madison, V, Cui, X, Thomas, A, Cheng, K.C. | | Deposit date: | 2009-06-16 | | Release date: | 2009-08-04 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Construction and characterization of a fully active PXR/SRC-1 tethered protein with increased stability
Protein Eng.Des.Sel., 21, 2008
|
|
5D0Q
 
 | | BamACDE complex, outer membrane beta-barrel assembly machinery (BAM) complex | | Descriptor: | Outer membrane protein assembly factor BamA, Outer membrane protein assembly factor BamC, Outer membrane protein assembly factor BamD, ... | | Authors: | Gu, Y, Paterson, N, Zeng, Y, Dong, H, Wang, W, Dong, C. | | Deposit date: | 2015-08-03 | | Release date: | 2016-03-09 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structural basis of outer membrane protein insertion by the BAM complex.
Nature, 531, 2016
|
|
2CNX
 
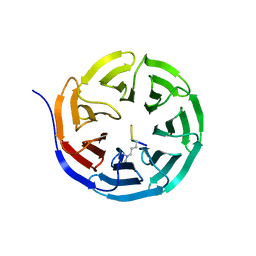 | | WDR5 and Histone H3 Lysine 4 dimethyl complex at 2.1 angstrom | | Descriptor: | HISTONE H3 DIMETHYL-LYSINE 4, WD-REPEAT PROTEIN 5 | | Authors: | Ruthenburg, A.J, Wang, W, Graybosch, D.M, Li, H, Allis, C.D, Patel, D.J, Verdine, G.L. | | Deposit date: | 2006-05-25 | | Release date: | 2006-07-03 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Histone H3 Recognition and Presentation by the Wdr5 Module of the Mll1 Complex
Nat.Struct.Mol.Biol., 13, 2006
|
|
2CO0
 
 | | WDR5 and unmodified Histone H3 complex at 2.25 Angstrom | | Descriptor: | HISTONE H3 DIMETHYL-LYSINE 4, WD-REPEAT PROTEIN 5 | | Authors: | Ruthenburg, A.J, Wang, W, Graybosch, D.M, Li, H, Allis, C.D, Patel, D.J, Verdine, G.L. | | Deposit date: | 2006-05-25 | | Release date: | 2006-07-03 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Histone H3 Recognition and Presentation by the Wdr5 Module of the Mll1 Complex
Nat.Struct.Mol.Biol., 13, 2006
|
|
4Q5W
 
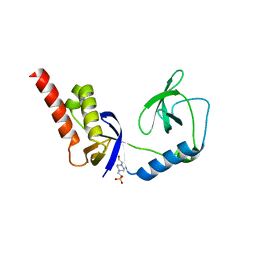 | | Crystal structure of extended-Tudor 9 of Drosophila melanogaster | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Maternal protein tudor | | Authors: | Ren, R, Liu, H, Wang, W, Wang, M, Yang, N, Dong, Y, Gong, W, Lehmann, R, Xu, R.M. | | Deposit date: | 2014-04-17 | | Release date: | 2014-05-21 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.801 Å) | | Cite: | Structure and domain organization of Drosophila Tudor
Cell Res., 24, 2014
|
|
2ARG
 
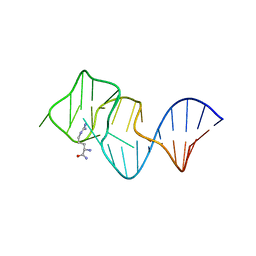 | | FORMATION OF AN AMINO ACID BINDING POCKET THROUGH ADAPTIVE ZIPPERING-UP OF A LARGE DNA HAIRPIN LOOP, NMR, 9 STRUCTURES | | Descriptor: | ARGININEAMIDE, DNA APTAMER [5'-D (*TP*GP*AP*CP*CP*AP*GP*GP*GP*CP*AP*AP*AP*CP*GP*GP*TP*AP* GP*GP*TP*GP*AP*GP*TP*GP*GP*TP*CP*A)-3'] | | Authors: | Lin, C.H, Wang, W, Jones, R.A, Patel, D.J. | | Deposit date: | 1998-08-19 | | Release date: | 1999-03-23 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Formation of an amino-acid-binding pocket through adaptive zippering-up of a large DNA hairpin loop.
Chem.Biol., 5, 1998
|
|
4Q5Y
 
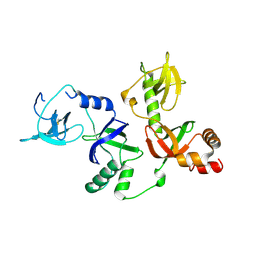 | | Crystal structure of extended-Tudor 10-11 of Drosophila melanogaster | | Descriptor: | Maternal protein tudor | | Authors: | Liu, H, Ren, R, Wang, W, Wang, M, Yang, N, Dong, Y, Gong, W, Lehmann, R, Xu, R.M. | | Deposit date: | 2014-04-18 | | Release date: | 2014-05-21 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure and domain organization of Drosophila Tudor
Cell Res., 24, 2014
|
|
4MGR
 
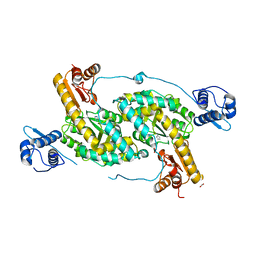 | | The crystal structure of Bacillus subtilis GabR, an autorepressor and PLP- and GABA-dependent transcriptional activator of gabT | | Descriptor: | ACETATE ION, HTH-type transcriptional regulatory protein GabR, IMIDAZOLE, ... | | Authors: | Wu, R, Edayathumangalam, R, Garcia, R, Wang, Y, Wang, W, Kreinbring, C.A, Bach, A, Liao, J, Stone, T, Terwilliger, T, Hoang, Q.Q, Belitsky, B.R, Petsko, G.A, Ringe, D, Liu, D. | | Deposit date: | 2013-08-28 | | Release date: | 2013-10-30 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Crystal structure of Bacillus subtilis GabR, an autorepressor and transcriptional activator of gabT.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4QVC
 
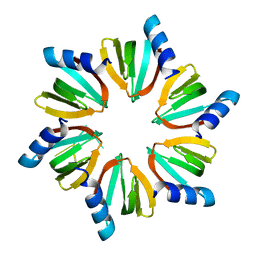 | | E.coli Hfq in complex with RNA Aus | | Descriptor: | RNA (5'-R(*AP*U*AP*AP*CP*UP*A)-3'), RNA-binding protein Hfq | | Authors: | Wang, L.J, Wang, W.W, Li, F.D, Wu, J.H, Gong, Q.G, Shi, Y.Y. | | Deposit date: | 2014-07-14 | | Release date: | 2015-05-27 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Structural insights into the recognition of the internal A-rich linker from OxyS sRNA by Escherichia coli Hfq
Nucleic Acids Res., 43, 2015
|
|
8E34
 
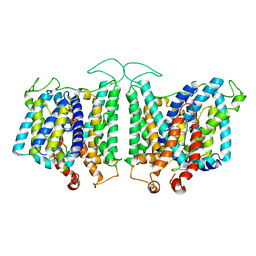 | | CryoEM structures of bAE1 captured in multiple states | | Descriptor: | Anion exchange protein | | Authors: | Zhekova, H.R, Wang, W.G, Jiang, J.S, Tsirulnikov, K, Muhammad-Khan, G.H, Azimov, R, Abuladze, N, Kao, L, Newman, D, Noskov, S.Y, Tieleman, P, Zhou, Z.H, Pushkin, A, Kurtz, I. | | Deposit date: | 2022-08-16 | | Release date: | 2023-01-25 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (6 Å) | | Cite: | CryoEM structures of anion exchanger 1 capture multiple states of inward- and outward-facing conformations.
Commun Biol, 5, 2022
|
|
1T6X
 
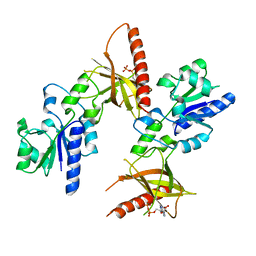 | | Crystal structure of ADP bound TM379 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, riboflavin kinase/FMN adenylyltransferase | | Authors: | Shin, D.H, Wang, W, Kim, R, Yokota, H, Kim, S.-H, Berkeley Structural Genomics Center (BSGC) | | Deposit date: | 2004-05-07 | | Release date: | 2004-08-10 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Crystal structure of ADP bound FAD synthetase
To be Published
|
|
1T6Z
 
 | | Crystal structure of riboflavin bound TM379 | | Descriptor: | RIBOFLAVIN, riboflavin kinase/FMN adenylyltransferase | | Authors: | Shin, D.H, Wang, W, Kim, R, Yokota, H, Kim, S.-H, Berkeley Structural Genomics Center (BSGC) | | Deposit date: | 2004-05-07 | | Release date: | 2004-08-10 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of ADP bound FAD synthetase
To be Published
|
|
5K4J
 
 | | Crystal Structure of CDK2 in complex with compound 22 | | Descriptor: | 1-[(1~{S})-1-(4-chloranyl-3-fluoranyl-phenyl)-2-oxidanyl-ethyl]-4-[2-[(2-methylpyrazol-3-yl)amino]pyrimidin-4-yl]pyridin-2-one, Cyclin-dependent kinase 2 | | Authors: | Yin, J, Wang, W. | | Deposit date: | 2016-05-20 | | Release date: | 2016-07-06 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Discovery of (S)-1-(1-(4-Chloro-3-fluorophenyl)-2-hydroxyethyl)-4-(2-((1-methyl-1H-pyrazol-5-yl)amino)pyrimidin-4-yl)pyridin-2(1H)-one (GDC-0994), an Extracellular Signal-Regulated Kinase 1/2 (ERK1/2) Inhibitor in Early Clinical Development.
J.Med.Chem., 59, 2016
|
|
6URC
 
 | | Crystal structure of IRE1a in complex with compound 18 | | Descriptor: | 2-chloro-N-(6-methyl-5-{[3-(2-{[(3S)-piperidin-3-yl]amino}pyrimidin-4-yl)pyridin-2-yl]oxy}naphthalen-1-yl)benzene-1-sulfonamide, GLYCEROL, Serine/threonine-protein kinase/endoribonuclease IRE1 | | Authors: | Wallweber, H.H, Wang, W. | | Deposit date: | 2019-10-23 | | Release date: | 2019-11-06 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Disruption of IRE1 alpha through its kinase domain attenuates multiple myeloma.
Proc. Natl. Acad. Sci. U.S.A., 116, 2019
|
|
5K4I
 
 | | Crystal Structure of ERK2 in complex with compound 22 | | Descriptor: | 1,2-ETHANEDIOL, 1-[(1~{S})-1-(4-chloranyl-3-fluoranyl-phenyl)-2-oxidanyl-ethyl]-4-[2-[(2-methylpyrazol-3-yl)amino]pyrimidin-4-yl]pyridin-2-one, Mitogen-activated protein kinase 1 | | Authors: | Yin, J, Wang, W. | | Deposit date: | 2016-05-20 | | Release date: | 2016-07-06 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Discovery of (S)-1-(1-(4-Chloro-3-fluorophenyl)-2-hydroxyethyl)-4-(2-((1-methyl-1H-pyrazol-5-yl)amino)pyrimidin-4-yl)pyridin-2(1H)-one (GDC-0994), an Extracellular Signal-Regulated Kinase 1/2 (ERK1/2) Inhibitor in Early Clinical Development.
J.Med.Chem., 59, 2016
|
|
8EEQ
 
 | | CryoEM structures of bAE1 captured in multiple states. | | Descriptor: | Anion exchange protein | | Authors: | Zhekova, H.R, Wang, W.G, Jiang, J.S, Tsirulnikov, K, Muhammad-Khan, G.H, Azimov, R, Abuladze, N, Kao, L, Newman, D, Noskov, S.Y, Tieleman, P, Zhou, Z.H, Pushkin, A, Kurtz, I. | | Deposit date: | 2022-09-07 | | Release date: | 2023-01-25 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (6.3 Å) | | Cite: | CryoEM structures of anion exchanger 1 capture multiple states of inward- and outward-facing conformations.
Commun Biol, 5, 2022
|
|
4B5X
 
 | | Crystal structures of divalent metal dependent pyruvate aldolase (HpaI), mutant D42A | | Descriptor: | 4-HYDROXY-2-OXO-HEPTANE-1,7-DIOATE ALDOLASE, GLYCEROL, PHOSPHATE ION | | Authors: | Coincon, M, Wang, W, Seah, S.Y.K, Sygusch, J. | | Deposit date: | 2012-08-07 | | Release date: | 2012-08-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of Reaction Intermediates in Pyruvate Class II Aldolase: Substrate Cleavage, Enolate Stabilization and Substrate Specificity
J.Biol.Chem., 287, 2012
|
|
