7ORR
 
 | | Non-structural protein 10 (nsp10) from SARS CoV-2 in complex with fragment VT00022 | | Descriptor: | 4-PHENYL-1H-IMIDAZOLE, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Talibov, V.O, Kozielski, F, Sele, C, Lou, J, Dong, D, Wang, Q, Shi, X, Nyblom, M, Rogstam, A, Krojer, T, Knecht, W, Fisher, S.Z. | | Deposit date: | 2021-06-06 | | Release date: | 2021-10-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Identification of fragments binding to SARS-CoV-2 nsp10 reveals ligand-binding sites in conserved interfaces between nsp10 and nsp14/nsp16.
Rsc Chem Biol, 3, 2022
|
|
7BOK
 
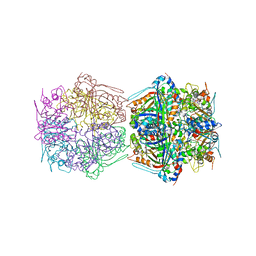 | | Cryo-EM structure of the encapsulated DyP-type peroxidase from Mycobacterium smegmatis | | Descriptor: | Dyp-type peroxidase, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Tang, Y.T, Mu, A, Gong, H.R, Wang, Q, Rao, Z.H. | | Deposit date: | 2020-03-19 | | Release date: | 2021-03-24 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Cryo-EM structure of Mycobacterium smegmatis DyP-loaded encapsulin.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7BOJ
 
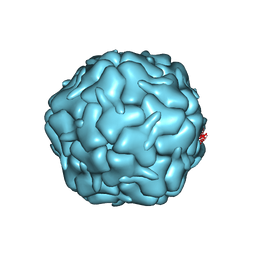 | | Cryo-EM structure of the encapsulin shell from Mycobacterium smegmatis | | Descriptor: | 29 kDa antigen Cfp29 | | Authors: | Tang, Y.T, Mu, A, Gong, H.R, Wang, Q, Rao, Z.H. | | Deposit date: | 2020-03-19 | | Release date: | 2021-03-24 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Cryo-EM structure of Mycobacterium smegmatis DyP-loaded encapsulin.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7ZL9
 
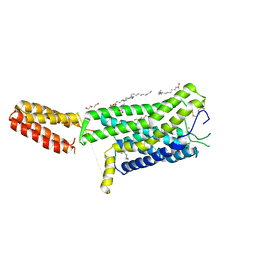 | | Crystal structure of human GPCR Niacin receptor (HCA2) | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, DI(HYDROXYETHYL)ETHER, Hydroxycarboxylic acid receptor 2,Soluble cytochrome b562, ... | | Authors: | Yang, Y, Kang, H.J, Gao, R.G, Wang, J.J, Han, G.W, FiBerto, J.F, Wu, L.J, Tong, J.H, Qu, L, Wu, Y.R, Pileski, R, Li, X.M, Zhang, X.C, Zhao, S.W, Kenakin, T, Wang, Q, Stevens, R.C, Peng, W, Roth, B.L, Rao, Z.H, Liu, Z.J. | | Deposit date: | 2022-04-14 | | Release date: | 2023-04-12 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural insights into the human niacin receptor HCA2-G i signalling complex.
Nat Commun, 14, 2023
|
|
7ZLY
 
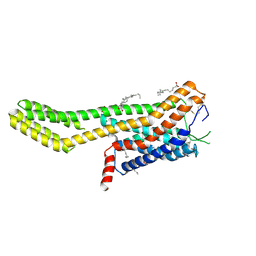 | | Crystal structure of human GPCR Niacin receptor (HCA2) expressed from Spodoptera frugiperda | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Hydroxycarboxylic acid receptor 2,Soluble cytochrome b562, OLEIC ACID | | Authors: | Yang, Y, Kang, H.J, Gao, R.G, Wang, J.J, FiBerto, J.F, Wu, L.J, Tong, J.H, Han, G.W, Qu, L, Wu, Y.R, Pileski, R, Li, X.M, Zhang, X.C, Zhao, S.W, Kenakin, T, Wang, Q, Stevens, R.C, Peng, W, Roth, B.L, Rao, Z.H, Liu, Z.J. | | Deposit date: | 2022-04-17 | | Release date: | 2023-04-12 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural insights into the human niacin receptor HCA2-G i signalling complex.
Nat Commun, 14, 2023
|
|
7YTL
 
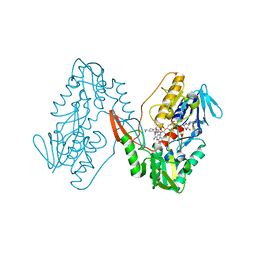 | | Structure of a oxidoreductase in complex with quinone | | Descriptor: | Apoptosis inducing factor mitochondria associated 2, FLAVIN-ADENINE DINUCLEOTIDE, UBIQUINONE-1 | | Authors: | Lv, Y, Sun, Q, Wang, Q, Zhu, D. | | Deposit date: | 2022-08-15 | | Release date: | 2023-08-23 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.62 Å) | | Cite: | Structural insights into FSP1 catalysis and ferroptosis inhibition.
Nat Commun, 14, 2023
|
|
1M8T
 
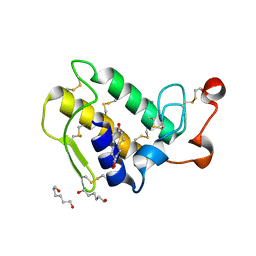 | | Structure of an acidic Phospholipase A2 from the venom of Ophiophagus hannah at 2.1 resolution from a hemihedrally twinned crystal form | | Descriptor: | CALCIUM ION, HEXANE-1,6-DIOL, Phospholipase a2 | | Authors: | Xu, S, Gu, L, Wang, Q, Shu, Y, Lin, Z. | | Deposit date: | 2002-07-26 | | Release date: | 2003-09-02 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of a king cobra phospholipase A2 determined from a hemihedrally twinned crystal.
Acta Crystallogr.,Sect.D, 59, 2003
|
|
4HU8
 
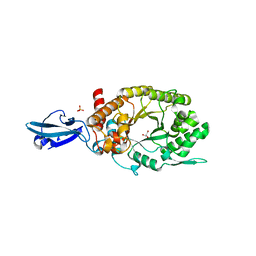 | | Crystal Structure of a Bacterial Ig-like Domain Containing GH10 Xylanase from Termite Gut | | Descriptor: | GH10 Xylanase, GLYCEROL, SULFATE ION | | Authors: | Han, Q, Liu, N, Robinson, H, Cao, L, Qian, C, Wang, Q, Xie, L, Ding, H, Wang, Q, Huang, Y, Li, J, Zhou, Z. | | Deposit date: | 2012-11-02 | | Release date: | 2013-09-18 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Biochemical characterization and crystal structure of a GH10 xylanase from termite gut bacteria reveal a novel structural feature and significance of its bacterial Ig-like domain.
Biotechnol.Bioeng., 110, 2013
|
|
5X58
 
 | | Prefusion structure of SARS-CoV spike glycoprotein, conformation 1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Yuan, Y, Cao, D, Zhang, Y, Ma, J, Qi, J, Wang, Q, Lu, G, Wu, Y, Yan, J, Shi, Y, Zhang, X, Gao, G.F. | | Deposit date: | 2017-02-15 | | Release date: | 2017-05-03 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Cryo-EM structures of MERS-CoV and SARS-CoV spike glycoproteins reveal the dynamic receptor binding domains
Nat Commun, 8, 2017
|
|
5X5B
 
 | | Prefusion structure of SARS-CoV spike glycoprotein, conformation 2 | | Descriptor: | Spike glycoprotein | | Authors: | Yuan, Y, Cao, D, Zhang, Y, Ma, J, Qi, J, Wang, Q, Lu, G, Wu, Y, Yan, J, Shi, Y, Zhang, X, Gao, G.F. | | Deposit date: | 2017-02-15 | | Release date: | 2017-05-03 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Cryo-EM structures of MERS-CoV and SARS-CoV spike glycoproteins reveal the dynamic receptor binding domains
Nat Commun, 8, 2017
|
|
5X59
 
 | | Prefusion structure of MERS-CoV spike glycoprotein, three-fold symmetry | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, S protein | | Authors: | Yuan, Y, Cao, D, Zhang, Y, Ma, J, Qi, J, Wang, Q, Lu, G, Wu, Y, Yan, J, Shi, Y, Zhang, X, Gao, G.F. | | Deposit date: | 2017-02-15 | | Release date: | 2017-05-03 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Cryo-EM structures of MERS-CoV and SARS-CoV spike glycoproteins reveal the dynamic receptor binding domains
Nat Commun, 8, 2017
|
|
5X5C
 
 | | Prefusion structure of MERS-CoV spike glycoprotein, conformation 1 | | Descriptor: | S protein | | Authors: | Yuan, Y, Cao, D, Zhang, Y, Ma, J, Qi, J, Wang, Q, Lu, G, Wu, Y, Yan, J, Shi, Y, Zhang, X, Gao, G.F. | | Deposit date: | 2017-02-15 | | Release date: | 2017-05-03 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Cryo-EM structures of MERS-CoV and SARS-CoV spike glycoproteins reveal the dynamic receptor binding domains
Nat Commun, 8, 2017
|
|
4RAZ
 
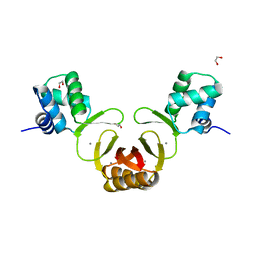 | | Crystal structure of Magnetospirillum gryphiswaldense MSR-1 holo-Fur | | Descriptor: | 1,2-ETHANEDIOL, DNA-binding transcriptional dual regulator of siderophore biosynthesis and transport(Fur family), MANGANESE (II) ION | | Authors: | Deng, Z, Wang, Q, Chen, Z. | | Deposit date: | 2014-09-12 | | Release date: | 2015-07-15 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Mechanistic insights into metal ion activation and operator recognition by the ferric uptake regulator.
Nat Commun, 6
|
|
6M71
 
 | | SARS-Cov-2 RNA-dependent RNA polymerase in complex with cofactors | | Descriptor: | Non-structural protein 7, Non-structural protein 8, RNA-directed RNA polymerase | | Authors: | Gao, Y, Yan, L, Huang, Y, Liu, F, Cao, L, Wang, T, Wang, Q, Lou, Z, Rao, Z. | | Deposit date: | 2020-03-16 | | Release date: | 2020-04-01 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structure of the RNA-dependent RNA polymerase from COVID-19 virus.
Science, 368, 2020
|
|
5X5F
 
 | | Prefusion structure of MERS-CoV spike glycoprotein, conformation 2 | | Descriptor: | S protein | | Authors: | Yuan, Y, Cao, D, Zhang, Y, Ma, J, Qi, J, Wang, Q, Lu, G, Wu, Y, Yan, J, Shi, Y, Zhang, X, Gao, G.F. | | Deposit date: | 2017-02-15 | | Release date: | 2017-05-03 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Cryo-EM structures of MERS-CoV and SARS-CoV spike glycoproteins reveal the dynamic receptor binding domains
Nat Commun, 8, 2017
|
|
6CXC
 
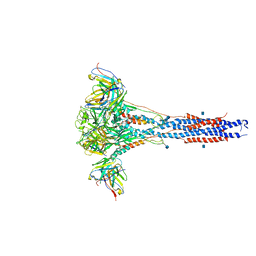 | | 3.9A Cryo-EM structure of murine antibody bound at a novel epitope of respiratory syncytial virus fusion protein | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Fusion glycoprotein F0, Envelope glycoprotein chimera, ... | | Authors: | Xie, Q, Wang, Z, Chen, X, Ni, F, Ma, J, Wang, Q. | | Deposit date: | 2018-04-02 | | Release date: | 2019-07-31 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structure basis of neutralization by a novel site II/IV antibody against respiratory syncytial virus fusion protein.
Plos One, 14, 2019
|
|
8YMK
 
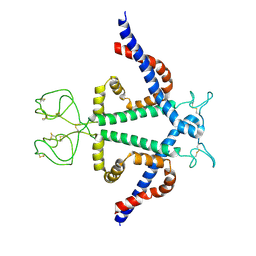 | | Localized reconstruction of Hepatitis B virus surface antigen dimer in the subviral particle with D2 symmetry from dataset A0 | | Descriptor: | Isoform S of Large envelope protein | | Authors: | Wang, T, Cao, L, Mu, A, Wang, Q, Rao, Z.H. | | Deposit date: | 2024-03-09 | | Release date: | 2024-09-18 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Inherent symmetry and flexibility in hepatitis B virus subviral particles.
Science, 385, 2024
|
|
8YMJ
 
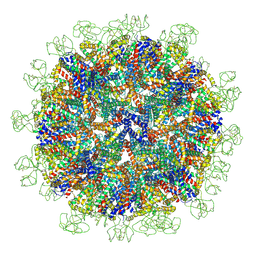 | | Cryo-EM structure of Hepatitis B virus surface antigen subviral particle with D2 symmetry | | Descriptor: | Isoform S of Large envelope protein | | Authors: | Wang, T, Cao, L, Mu, A, Wang, Q, Rao, Z.H. | | Deposit date: | 2024-03-09 | | Release date: | 2024-09-18 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (6.6 Å) | | Cite: | Inherent symmetry and flexibility in hepatitis B virus subviral particles.
Science, 385, 2024
|
|
7T90
 
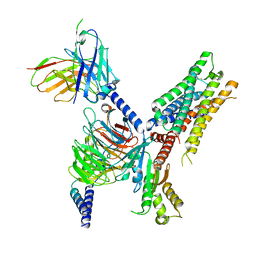 | | Cryo-EM structure of ACh-bound M2R-Go signaling complex in S2 state | | Descriptor: | ACETYLCHOLINE, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Xu, J, Wang, Q, Du, Y, Kobilka, B.K. | | Deposit date: | 2021-12-17 | | Release date: | 2023-01-25 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.32 Å) | | Cite: | Structural and dynamic insights into supra-physiological activation and allosteric modulation of a muscarinic acetylcholine receptor.
Nat Commun, 14, 2023
|
|
7T8X
 
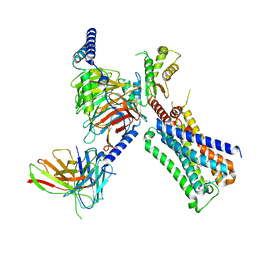 | | Cryo-EM structure of ACh-bound M2R-Go signaling complex in S1 state | | Descriptor: | ACETYLCHOLINE, Antibody fragment, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Xu, J, Wang, Q, Du, Y, Kobilka, B.K. | | Deposit date: | 2021-12-17 | | Release date: | 2023-01-25 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.21 Å) | | Cite: | Structural and dynamic insights into supra-physiological activation and allosteric modulation of a muscarinic acetylcholine receptor.
Nat Commun, 14, 2023
|
|
7T94
 
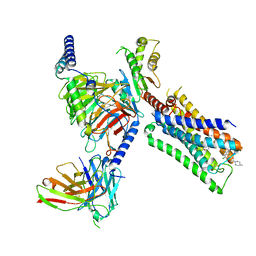 | | Cryo-EM structure of S1 state ACh-bound M2R-Go signaling complex with a PAM | | Descriptor: | 3-amino-5-chloro-N-cyclopropyl-4-methyl-6-[2-(4-methylpiperazin-1-yl)-2-oxoethoxy]thieno[2,3-b]pyridine-2-carboxamide, ACETYLCHOLINE, Antibody fragment, ... | | Authors: | Xu, J, Wang, Q, Du, Y, Kobilka, B.K. | | Deposit date: | 2021-12-17 | | Release date: | 2023-01-25 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.16 Å) | | Cite: | Structural and dynamic insights into supra-physiological activation and allosteric modulation of a muscarinic acetylcholine receptor.
Nat Commun, 14, 2023
|
|
7T96
 
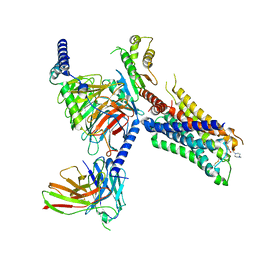 | | Cryo-EM structure of S2 state ACh-bound M2R-Go signaling complex with a PAM | | Descriptor: | 3-amino-5-chloro-N-cyclopropyl-4-methyl-6-[2-(4-methylpiperazin-1-yl)-2-oxoethoxy]thieno[2,3-b]pyridine-2-carboxamide, ACETYLCHOLINE, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Xu, J, Wang, Q, Du, Y, Kobilka, B.K. | | Deposit date: | 2021-12-18 | | Release date: | 2023-01-25 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.22 Å) | | Cite: | Structural and dynamic insights into supra-physiological activation and allosteric modulation of a muscarinic acetylcholine receptor.
Nat Commun, 14, 2023
|
|
3N3Y
 
 | | Crystal structure of Thymidylate Synthase X (ThyX) from Helicobacter pylori with FAD and dUMP at 2.31A resolution | | Descriptor: | 2'-DEOXYURIDINE 5'-MONOPHOSPHATE, FLAVIN-ADENINE DINUCLEOTIDE, Thymidylate synthase thyX | | Authors: | Wang, K, Wang, Q, Chen, J, Chen, L, Jiang, H, Shen, X. | | Deposit date: | 2010-05-21 | | Release date: | 2011-05-25 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.307 Å) | | Cite: | Structure, Enzymatic Characterization and Inhibitor Discovery of Thymidylate Synthase X (ThyX) from Helicobacter pylori Strain SS1
To be Published
|
|
7BX8
 
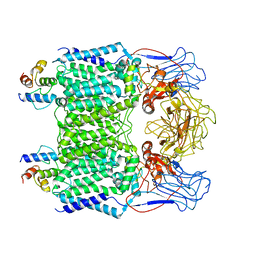 | | Mycobacterium smegmatis arabinosyltransferase complex EmbB2-AcpM2 in symmetric "resting state" | | Descriptor: | Integral membrane indolylacetylinositol arabinosyltransferase EmbB, Meromycolate extension acyl carrier protein | | Authors: | Gao, R.G, Zhang, L, Wang, Q, Rao, Z.H. | | Deposit date: | 2020-04-17 | | Release date: | 2020-05-27 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Cryo-EM snapshots of mycobacterial arabinosyltransferase complex EmbB2-AcpM2.
Protein Cell, 11, 2020
|
|
7BWR
 
 | | Mycobacterium smegmatis arabinosyltransferase complex EmbB2-AcpM2 in substrate DPA bound asymmetric "active state" | | Descriptor: | CALCIUM ION, Integral membrane indolylacetylinositol arabinosyltransferase EmbB, Meromycolate extension acyl carrier protein, ... | | Authors: | Gao, R.G, Zhang, L, Wang, Q, Rao, Z.H. | | Deposit date: | 2020-04-15 | | Release date: | 2020-05-27 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Cryo-EM snapshots of mycobacterial arabinosyltransferase complex EmbB2-AcpM2.
Protein Cell, 11, 2020
|
|
