7WNH
 
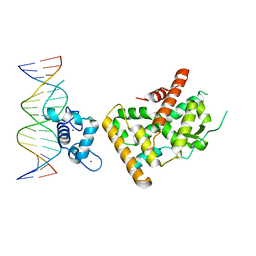 | | Crystal structure of Nurr1 binding to NBRE | | Descriptor: | DNA (5'-D(*CP*CP*GP*AP*AP*AP*AP*GP*GP*TP*CP*AP*TP*GP*CP*G)-3'), DNA (5'-D(*CP*GP*CP*AP*TP*GP*AP*CP*CP*TP*TP*TP*TP*CP*GP*G)-3'), Nuclear receptor subfamily 4 group A member 2, ... | | Authors: | Zhao, M, Xu, T, Wang, N, Guo, Y, Liu, J. | | Deposit date: | 2022-01-18 | | Release date: | 2023-01-25 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Integrative analysis reveals structural basis for transcription activation of Nurr1 and Nurr1-RXR alpha heterodimer.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7DFL
 
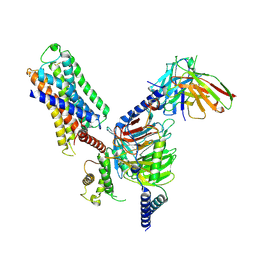 | | Cryo-EM structure of histamine H1 receptor Gq complex | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(q) subunit alpha, ... | | Authors: | He, Y, Xia, R, Wang, N, Xu, Z. | | Deposit date: | 2020-11-09 | | Release date: | 2021-03-31 | | Last modified: | 2021-04-21 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Cryo-EM structure of the human histamine H 1 receptor/G q complex.
Nat Commun, 12, 2021
|
|
7CRZ
 
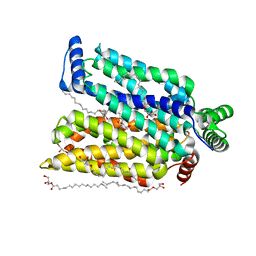 | | Crystal structure of human glucose transporter GLUT3 bound with C3361 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (2S,3R,4S,5R,6R)-6-(hydroxymethyl)-4-undec-10-enoxy-oxane-2,3,5-triol, Solute carrier family 2, ... | | Authors: | Yuan, Y.Y, Zhang, S, Wang, N, Jiang, X, Yan, N. | | Deposit date: | 2020-08-14 | | Release date: | 2021-01-13 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Orthosteric-allosteric dual inhibitors of PfHT1 as selective antimalarial agents.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
3AZX
 
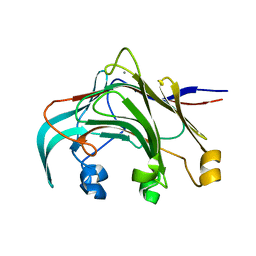 | | Crystal structure of the laminarinase catalytic domain from Thermotoga maritima MSB8 | | Descriptor: | CALCIUM ION, Laminarinase | | Authors: | Jeng, W.Y, Wang, N.C, Wang, A.H.J. | | Deposit date: | 2011-06-03 | | Release date: | 2011-11-23 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structures of the laminarinase catalytic domain from Thermotoga maritima MSB8 in complex with inhibitors: essential residues for beta-1,3 and beta-1,4 glucan selection.
J.Biol.Chem., 286, 2011
|
|
3B01
 
 | | Crystal structure of the laminarinase catalytic domain from Thermotoga maritima MSB8 | | Descriptor: | CALCIUM ION, CHLORIDE ION, Laminarinase, ... | | Authors: | Jeng, W.Y, Wang, N.C, Wang, A.H.J. | | Deposit date: | 2011-06-03 | | Release date: | 2011-11-23 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Crystal structures of the laminarinase catalytic domain from Thermotoga maritima MSB8 in complex with inhibitors: essential residues for beta-1,3 and beta-1,4 glucan selection.
J.Biol.Chem., 286, 2011
|
|
3AZZ
 
 | | Crystal structure of the laminarinase catalytic domain from Thermotoga maritima MSB8 in complex with gluconolactone | | Descriptor: | CALCIUM ION, D-glucono-1,5-lactone, Laminarinase, ... | | Authors: | Jeng, W.Y, Wang, N.C, Wang, A.H.J. | | Deposit date: | 2011-06-03 | | Release date: | 2011-11-23 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Crystal structures of the laminarinase catalytic domain from Thermotoga maritima MSB8 in complex with inhibitors: essential residues for beta-1,3 and beta-1,4 glucan selection.
J.Biol.Chem., 286, 2011
|
|
3AZY
 
 | | Crystal structure of the laminarinase catalytic domain from Thermotoga maritima MSB8 | | Descriptor: | CALCIUM ION, Laminarinase, SULFATE ION | | Authors: | Jeng, W.Y, Wang, N.C, Wang, A.H.J. | | Deposit date: | 2011-06-03 | | Release date: | 2011-11-23 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal structures of the laminarinase catalytic domain from Thermotoga maritima MSB8 in complex with inhibitors: essential residues for beta-1,3 and beta-1,4 glucan selection.
J.Biol.Chem., 286, 2011
|
|
3B00
 
 | | Crystal structure of the laminarinase catalytic domain from Thermotoga maritima MSB8 in complex with cetyltrimethylammonium bromide | | Descriptor: | CALCIUM ION, CETYL-TRIMETHYL-AMMONIUM, Laminarinase | | Authors: | Jeng, W.Y, Wang, N.C, Wang, A.H.J. | | Deposit date: | 2011-06-03 | | Release date: | 2011-11-23 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Crystal structures of the laminarinase catalytic domain from Thermotoga maritima MSB8 in complex with inhibitors: essential residues for beta-1,3 and beta-1,4 glucan selection.
J.Biol.Chem., 286, 2011
|
|
2ZY5
 
 | | R487A mutant of L-aspartate beta-decarboxylase | | Descriptor: | L-aspartate beta-decarboxylase, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Chen, H.-J, Ko, T.-P, Lee, C.-Y, Wang, N.-C, Wang, A.H.-J. | | Deposit date: | 2009-01-13 | | Release date: | 2009-01-27 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structure, Assembly, and Mechanism of a PLP-Dependent Dodecameric l-Aspartate beta-Decarboxylase
Structure, 17, 2009
|
|
2ZY2
 
 | | dodecameric L-aspartate beta-decarboxylase | | Descriptor: | L-aspartate 4-carboxylyase, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Chen, H.-J, Ko, T.-P, Lee, C.-Y, Wang, N.-C, Wang, A.H.-J. | | Deposit date: | 2009-01-13 | | Release date: | 2009-01-27 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structure, Assembly, and Mechanism of a PLP-Dependent Dodecameric l-Aspartate beta-Decarboxylase
Structure, 17, 2009
|
|
2ZY3
 
 | | dodecameric L-aspartate beta-decarboxylase | | Descriptor: | L-aspartate beta-decarboxylase, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Chen, H.-J, Ko, T.-P, Lee, C.-Y, Wang, N.-C, Wang, A.H.-J. | | Deposit date: | 2009-01-13 | | Release date: | 2009-01-27 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure, Assembly, and Mechanism of a PLP-Dependent Dodecameric l-Aspartate beta-Decarboxylase
Structure, 17, 2009
|
|
2ZY4
 
 | | dodecameric L-aspartate beta-decarboxylase | | Descriptor: | CHLORIDE ION, L-aspartate beta-decarboxylase, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Chen, H.-J, Ko, T.-P, Lee, C.-Y, Wang, N.-C, Wang, A.H.-J. | | Deposit date: | 2009-01-13 | | Release date: | 2009-01-27 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure, Assembly, and Mechanism of a PLP-Dependent Dodecameric l-Aspartate beta-Decarboxylase
Structure, 17, 2009
|
|
2KAW
 
 | | NMR structure of the mDvl1 PDZ domain in complex with its inhibitor | | Descriptor: | Segment polarity protein dishevelled homolog DVL-1, [(1Z)-5-fluoro-2-methyl-1-{4-[methylsulfinyl]benzylidene}-1H-inden-3-yl]acetic acid | | Authors: | Lee, H.J, Shao, Y, Wang, N.X, Shi, D.L, Zheng, J.J. | | Deposit date: | 2008-11-17 | | Release date: | 2009-09-15 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Sulindac inhibits canonical Wnt signaling by blocking the PDZ domain of the protein Dishevelled.
Angew.Chem.Int.Ed.Engl., 48, 2009
|
|
