8JCV
 
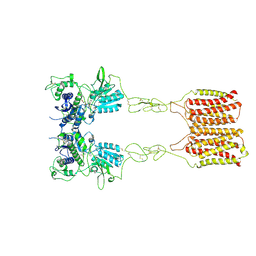 | | Cryo-EM structure of mGlu2-mGlu3 heterodimer in presence of LY341495 (dimerization mode II) | | Descriptor: | 2-[(1S,2S)-2-carboxycyclopropyl]-3-(9H-xanthen-9-yl)-D-alanine, 2-acetamido-2-deoxy-beta-D-glucopyranose, Metabotropic glutamate receptor 2,Peptidyl-prolyl cis-trans isomerase FKBP1A, ... | | Authors: | Wang, X, Wang, M, Xu, T, Feng, Y, Han, S, Zhao, Q, Wu, B. | | Deposit date: | 2023-05-12 | | Release date: | 2023-06-21 | | Last modified: | 2023-10-18 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural insights into dimerization and activation of the mGlu2-mGlu3 and mGlu2-mGlu4 heterodimers.
Cell Res., 33, 2023
|
|
8JD6
 
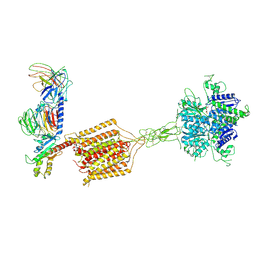 | | Cryo-EM structure of Gi1-bound metabotropic glutamate receptor mGlu4 | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(i) subunit alpha-3, ... | | Authors: | Wang, X, Wang, M, Xu, T, Feng, Y, Han, S, Lin, S, Zhao, Q, Wu, B. | | Deposit date: | 2023-05-12 | | Release date: | 2023-06-21 | | Last modified: | 2023-10-18 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural insights into dimerization and activation of the mGlu2-mGlu3 and mGlu2-mGlu4 heterodimers.
Cell Res., 33, 2023
|
|
8JCY
 
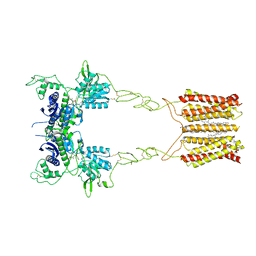 | | Cryo-EM structure of mGlu2-mGlu3 heterodimer in presence of LY341495, NAM563, and LY2389575 (dimerization mode I) | | Descriptor: | 2-[(1S,2S)-2-carboxycyclopropyl]-3-(9H-xanthen-9-yl)-D-alanine, 2-acetamido-2-deoxy-beta-D-glucopyranose, CHOLESTEROL, ... | | Authors: | Wang, X, Wang, M, Xu, T, Feng, Y, Han, S, Lin, S, Zhao, Q, Wu, B. | | Deposit date: | 2023-05-12 | | Release date: | 2023-06-21 | | Last modified: | 2023-10-18 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural insights into dimerization and activation of the mGlu2-mGlu3 and mGlu2-mGlu4 heterodimers.
Cell Res., 33, 2023
|
|
8JD0
 
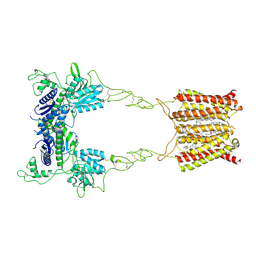 | | Cryo-EM structure of mGlu2-mGlu3 heterodimer in presence of NAM563 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(1-methylpyrazol-4-yl)-7-[[(2~{S})-2-(trifluoromethyl)morpholin-4-yl]methyl]quinoline-2-carboxamide, CHOLESTEROL, ... | | Authors: | Wang, X, Wang, M, Xu, T, Feng, Y, Han, S, Lin, S, Zhao, Q, Wu, B. | | Deposit date: | 2023-05-12 | | Release date: | 2023-06-21 | | Last modified: | 2023-10-18 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural insights into dimerization and activation of the mGlu2-mGlu3 and mGlu2-mGlu4 heterodimers.
Cell Res., 33, 2023
|
|
8JCU
 
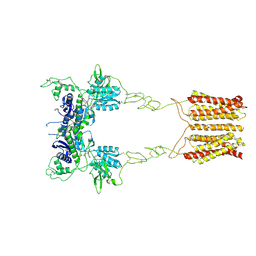 | | Cryo-EM structure of mGlu2-mGlu3 heterodimer in presence of LY341495 (dimerization mode I) | | Descriptor: | 2-[(1S,2S)-2-carboxycyclopropyl]-3-(9H-xanthen-9-yl)-D-alanine, 2-acetamido-2-deoxy-beta-D-glucopyranose, Metabotropic glutamate receptor 2,Peptidyl-prolyl cis-trans isomerase FKBP1A, ... | | Authors: | Wang, X, Wang, M, Xu, T, Feng, Y, Zhao, Q, Wu, B. | | Deposit date: | 2023-05-12 | | Release date: | 2023-06-21 | | Last modified: | 2023-10-18 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structural insights into dimerization and activation of the mGlu2-mGlu3 and mGlu2-mGlu4 heterodimers.
Cell Res., 33, 2023
|
|
8JD5
 
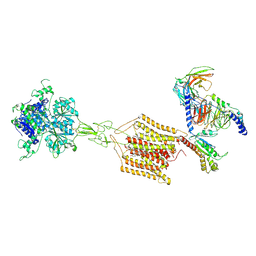 | | Cryo-EM structure of Gi1-bound mGlu2-mGlu4 heterodimer | | Descriptor: | 1-butyl-3-chloranyl-4-(4-phenylpiperidin-1-yl)pyridin-2-one, 2-acetamido-2-deoxy-beta-D-glucopyranose, 5-methyl-N-(4-methylpyrimidin-2-yl)-4-(1H-pyrazol-4-yl)-1,3-thiazol-2-amine, ... | | Authors: | Wang, X, Wang, M, Xu, T, Feng, Y, Han, S, Zhao, Q, Wu, B. | | Deposit date: | 2023-05-12 | | Release date: | 2023-06-21 | | Last modified: | 2023-10-18 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural insights into dimerization and activation of the mGlu2-mGlu3 and mGlu2-mGlu4 heterodimers.
Cell Res., 33, 2023
|
|
8JCX
 
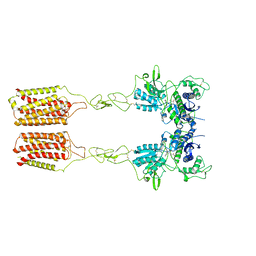 | | Cryo-EM structure of mGlu2-mGlu3 heterodimer in presence of LY341495 and NAM563 (dimerization mode II) | | Descriptor: | 2-[(1S,2S)-2-carboxycyclopropyl]-3-(9H-xanthen-9-yl)-D-alanine, 2-acetamido-2-deoxy-beta-D-glucopyranose, Metabotropic glutamate receptor 2,Peptidyl-prolyl cis-trans isomerase FKBP1A, ... | | Authors: | Wang, X, Wang, M, Xu, T, Feng, Y, Han, S, Lin, S, Zhao, Q, Wu, B. | | Deposit date: | 2023-05-12 | | Release date: | 2023-06-21 | | Last modified: | 2023-10-18 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural insights into dimerization and activation of the mGlu2-mGlu3 and mGlu2-mGlu4 heterodimers.
Cell Res., 33, 2023
|
|
8JD4
 
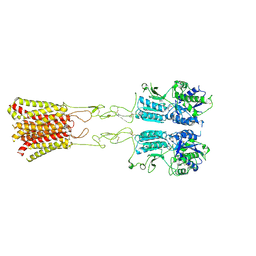 | | Cryo-EM structure of G protein-free mGlu2-mGlu4 heterodimer in Acc state | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GLUTAMIC ACID, Metabotropic glutamate receptor 2,Peptidyl-prolyl cis-trans isomerase FKBP1A, ... | | Authors: | Wang, X, Wang, M, Xu, T, Feng, Y, Han, S, Lin, S, Zhao, Q, Wu, B. | | Deposit date: | 2023-05-12 | | Release date: | 2023-06-21 | | Last modified: | 2023-10-18 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural insights into dimerization and activation of the mGlu2-mGlu3 and mGlu2-mGlu4 heterodimers.
Cell Res., 33, 2023
|
|
8JD3
 
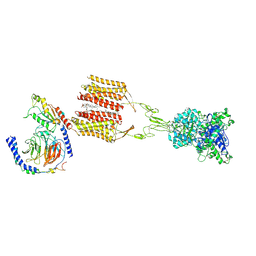 | | Cryo-EM structure of Gi1-bound mGlu2-mGlu3 heterodimer | | Descriptor: | 1-butyl-3-chloranyl-4-(4-phenylpiperidin-1-yl)pyridin-2-one, CHOLESTEROL, DI-PALMITOYL-3-SN-PHOSPHATIDYLETHANOLAMINE, ... | | Authors: | Wang, X, Wang, M, Xu, T, Feng, Y, Han, S, Lin, S, Zhao, Q, Wu, B. | | Deposit date: | 2023-05-12 | | Release date: | 2023-06-21 | | Last modified: | 2023-10-18 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural insights into dimerization and activation of the mGlu2-mGlu3 and mGlu2-mGlu4 heterodimers.
Cell Res., 33, 2023
|
|
4IYG
 
 | |
5H52
 
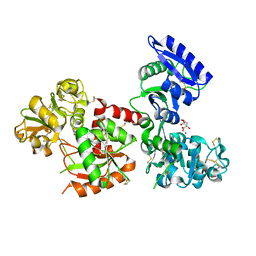 | | Structure of Titanium-bound human serum transferrin | | Descriptor: | CITRIC ACID, MALONATE ION, Serotransferrin, ... | | Authors: | Curtin, J.P, Wang, M, Sun, H. | | Deposit date: | 2016-11-04 | | Release date: | 2017-11-08 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The role of citrate, lactate and transferrin in determining titanium release from surgical devices into human serum.
J. Biol. Inorg. Chem., 23, 2018
|
|
5FM0
 
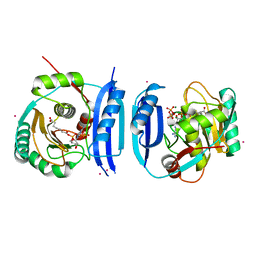 | | Crystal structure of the 6-carboxyhexanoate-CoA ligase (BioW)from Bacillus subtilis (PtCl4 derivative) | | Descriptor: | 6-CARBOXYHEXANOATE--COA LIGASE, MAGNESIUM ION, PIMELOYL-AMP, ... | | Authors: | Moynie, L, Wang, M, Campopiano, D.J, Naismith, J.H. | | Deposit date: | 2015-10-29 | | Release date: | 2016-11-16 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | Using the pimeloyl-CoA synthetase adenylation fold to synthesize fatty acid thioesters.
Nat. Chem. Biol., 13, 2017
|
|
5G1F
 
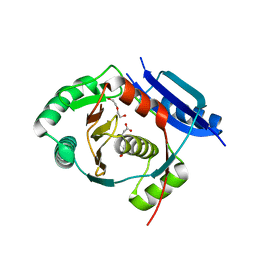 | | Crystal structure of the 6-carboxyhexanoate-CoA ligase (BioW)from Bacillus subtilis in complex with coenzyme A | | Descriptor: | 6-CARBOXYHEXANOATE-COA LIGASE, COENZYME A, NONAETHYLENE GLYCOL, ... | | Authors: | Moynie, L, Wang, M, Campopiano, D.J, Naismith, J.H. | | Deposit date: | 2016-03-25 | | Release date: | 2017-03-08 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | The Structure of the 6-Carboxyhexanoate-Coa Ligase from Bacillus Subtilis
To be Published
|
|
4IMB
 
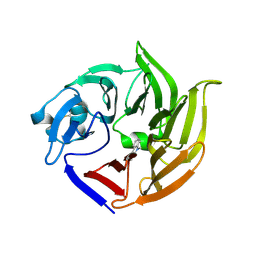 | |
4J12
 
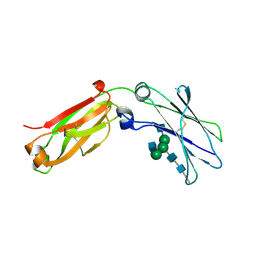 | | monomeric Fc | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, human Fc fragment | | Authors: | Ishino, T, Wang, M, Mosyak, L, Tam, A, Duan, W, Svenson, K, Joyce, A, O'Hara, D, Lin, L, Somers, W, Kriz, R. | | Deposit date: | 2013-01-31 | | Release date: | 2013-05-01 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Engineering a Monomeric Fc Domain Modality by N-Glycosylation for the Half-life Extension of Biotherapeutics.
J.Biol.Chem., 288, 2013
|
|
3GYR
 
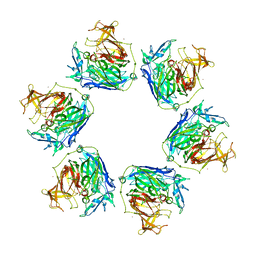 | | Structure of Phenoxazinone synthase from Streptomyces antibioticus reveals a new type 2 copper center. | | Descriptor: | COPPER (II) ION, CU-O-CU LINKAGE, GLYCEROL, ... | | Authors: | Smith, A.W, Camara-Artigas, A, Wang, M, Francisco, W.A, Allen, J.P. | | Deposit date: | 2009-04-05 | | Release date: | 2009-04-28 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of Phenoxazinone Synthase from Streptomyces Antibioticus Reveals a New Type 2 Copper Center.
Biochemistry, 45, 2006
|
|
7CGC
 
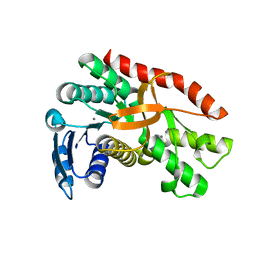 | |
6LSH
 
 | | Crystal structure of the enterovirus 71 polymerase elongation complex (C2S6M form) | | Descriptor: | Genome polyprotein, RNA (35-MER), RNA (5'-R(*UP*GP*UP*UP*CP*GP*AP*CP*GP*AP*GP*AP*GP*AP*GP*AP*CP*C)-3'), ... | | Authors: | Li, R, Wang, M, Jing, X, Gong, P. | | Deposit date: | 2020-01-17 | | Release date: | 2020-04-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.231 Å) | | Cite: | Stringent control of the RNA-dependent RNA polymerase translocation revealed by multiple intermediate structures.
Nat Commun, 11, 2020
|
|
3MDD
 
 | |
6LSG
 
 | | Crystal structure of the enterovirus 71 polymerase elongation complex (C0S6M form) | | Descriptor: | Genome polyprotein, RNA (35-MER), RNA (5'-R(*UP*GP*UP*UP*CP*GP*AP*CP*GP*AP*GP*AP*GP*AP*GP*A)-3'), ... | | Authors: | Li, R, Wang, M, Jing, X, Gong, P. | | Deposit date: | 2020-01-17 | | Release date: | 2020-04-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Stringent control of the RNA-dependent RNA polymerase translocation revealed by multiple intermediate structures.
Nat Commun, 11, 2020
|
|
6M40
 
 | | Crystal structure of the NS3-like helicase from Alongshan virus | | Descriptor: | NS3-like protein | | Authors: | Gao, X.P, Zhu, K.X, Chen, P, Wojdyla, J.A, Wang, M, Cui, S. | | Deposit date: | 2020-03-05 | | Release date: | 2020-04-08 | | Last modified: | 2020-06-03 | | Method: | X-RAY DIFFRACTION (2.89 Å) | | Cite: | Crystal structure of the NS3-like helicase from Alongshan virus.
Iucrj, 7, 2020
|
|
3MDE
 
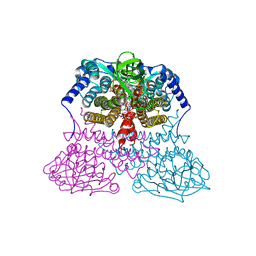 | |
7VAB
 
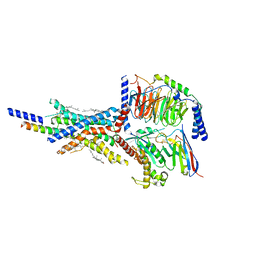 | | Cryo-EM structure of the non-acylated tirzepatide (LY3298176)-bound human GIPR-Gs complex | | Descriptor: | CHOLESTEROL, Gastric inhibitory polypeptide receptor,Gastric inhibitory polypeptide receptor,Gastric inhibitory polypeptide receptor,human glucose-dependent insulinotropic polypeptide receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Zhao, F.H, Zhou, Q.T, Cong, Z.T, Hang, K.N, Zou, X.Y, Zhang, C, Chen, Y, Dai, A.T, Liang, A.Y, Ming, Q.Q, Wang, M, Chen, L.N, Xu, P.Y, Chang, R.L, Feng, W.B, Xia, T, Zhang, Y, Wu, B.L, Yang, D.H, Zhao, L.H, Xu, H.E, Wang, M.W. | | Deposit date: | 2021-08-28 | | Release date: | 2022-03-02 | | Last modified: | 2022-03-16 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural insights into multiplexed pharmacological actions of tirzepatide and peptide 20 at the GIP, GLP-1 or glucagon receptors.
Nat Commun, 13, 2022
|
|
7VBH
 
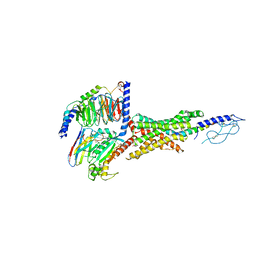 | | Cryo-EM structure of the GIPR/GLP-1R/GCGR triagonist peptide 20-bound human GLP-1R-Gs complex | | Descriptor: | Glucagon-like peptide 1 receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Zhao, F.H, Zhou, Q.T, Cong, Z.T, Hang, K.N, Zou, X.Y, Zhang, C, Chen, Y, Dai, A.T, Liang, A.Y, Ming, Q.Q, Wang, M, Chen, L.N, Xu, P.Y, Chang, R.L, Feng, W.B, Xia, T, Zhang, Y, Wu, B.L, Yang, D.H, Zhao, L.H, Xu, H.E, Wang, M.W. | | Deposit date: | 2021-08-31 | | Release date: | 2022-04-06 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural insights into multiplexed pharmacological actions of tirzepatide and peptide 20 at the GIP, GLP-1 or glucagon receptors.
Nat Commun, 13, 2022
|
|
7CGD
 
 | | Silver-bound E.coli malate dehydrogenase | | Descriptor: | Malate dehydrogenase, SILVER ION | | Authors: | Wang, H, Wang, M, Sun, H. | | Deposit date: | 2020-07-01 | | Release date: | 2020-09-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Atomic differentiation of silver binding preference in protein targets: Escherichia coli malate dehydrogenase as a paradigm.
Chem Sci, 11, 2020
|
|
