8YOO
 
 | | Cryo-EM structure of the human 80S ribosome with 100 um Tigecycline | | Descriptor: | 18S rRNA, 28S rRNA, 40S ribosomal protein S10, ... | | Authors: | Li, X, Wang, M, Denk, T, Cheng, J. | | Deposit date: | 2024-03-13 | | Release date: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (2 Å) | | Cite: | Structural basis for differential inhibition of eukaryotic ribosomes by tigecycline.
Nat Commun, 15, 2024
|
|
6RYO
 
 | | Bacterial membrane enzyme structure by the in meso method at 1.9 A resolution | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 2-(2-(2-(2-(2-(2-ETHOXYETHOXY)ETHOXY)ETHOXY)ETHOXY)ETHOXY)ETHANOL, GLYCEROL, ... | | Authors: | Huang, C.Y, Olatunji, S, Bailey, J, Yu, X, Olieric, V, Wang, M, Caffrey, M. | | Deposit date: | 2019-06-11 | | Release date: | 2020-01-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.924 Å) | | Cite: | Structures of lipoprotein signal peptidase II from Staphylococcus aureus complexed with antibiotics globomycin and myxovirescin.
Nat Commun, 11, 2020
|
|
5DQU
 
 | | Crystal Structure of Cas-DNA-10 complex | | Descriptor: | CRISPR-associated endonuclease Cas1, CRISPR-associated endoribonuclease Cas2, DNA (5'-D(*GP*AP*GP*TP*CP*GP*AP*TP*GP*CP*TP*TP*TP*TP*T)-3'), ... | | Authors: | Wang, J, Li, J, Zhao, H, Sheng, G, Wang, M, Yin, M, Wang, Y. | | Deposit date: | 2015-09-15 | | Release date: | 2015-11-11 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (4.5 Å) | | Cite: | Structural and Mechanistic Basis of PAM-Dependent Spacer Acquisition in CRISPR-Cas Systems.
Cell, 163, 2015
|
|
6G8A
 
 | | Lysozyme solved by Native SAD from a dataset collected in 5 seconds at 1 A wavelength with JUNGFRAU detector | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Lysozyme C, ... | | Authors: | Leonarski, F, Olieric, V, Vera, L, Redford, S, Wang, M. | | Deposit date: | 2018-04-08 | | Release date: | 2018-08-01 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.143 Å) | | Cite: | Fast and accurate data collection for macromolecular crystallography using the JUNGFRAU detector.
Nat. Methods, 15, 2018
|
|
6FMX
 
 | | IMISX-EP of W-PgpB | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Phosphatidylglycerophosphatase B, TUNGSTATE(VI)ION | | Authors: | Huang, C.-Y, Olieric, V, Howe, N, Warshamanage, R, Weinert, T, Panepucci, E, Vogeley, L, Basu, S, Diederichs, K, Caffrey, M, Wang, M. | | Deposit date: | 2018-02-02 | | Release date: | 2018-09-05 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | In situ serial crystallography for rapid de novo membrane protein structure determination.
Commun Biol, 1, 2018
|
|
6RYP
 
 | | Bacterial membrane enzyme structure by the in meso method at 2.3 A resolution | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, DI(HYDROXYETHYL)ETHER, Lipoprotein signal peptidase, ... | | Authors: | Huang, C.Y, Olatunji, S, Bailey, J, Yu, X, Olieric, V, Wang, M, Caffrey, M. | | Deposit date: | 2019-06-11 | | Release date: | 2020-01-15 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structures of lipoprotein signal peptidase II from Staphylococcus aureus complexed with antibiotics globomycin and myxovirescin.
Nat Commun, 11, 2020
|
|
8YW3
 
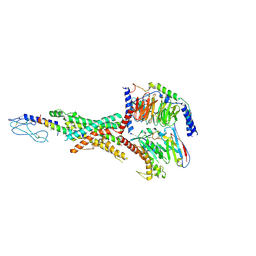 | | Cryo-EM structure of the retatrutide-bound human GLP-1R-Gs complex | | Descriptor: | Glucagon-like peptide 1 receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Li, W.Z, Zhou, Q.T, Cong, Z.T, Yuan, Q.N, Li, W.X, Zhao, F.H, Xu, H.E, Zhao, L.H, Yang, D.H, Wang, M.W. | | Deposit date: | 2024-03-29 | | Release date: | 2024-09-18 | | Last modified: | 2024-09-25 | | Method: | ELECTRON MICROSCOPY (2.68 Å) | | Cite: | Structural insights into the triple agonism at GLP-1R, GIPR and GCGR manifested by retatrutide.
Cell Discov, 10, 2024
|
|
8YW4
 
 | | Cryo-EM structure of the retatrutide-bound human GIPR-Gs complex | | Descriptor: | Gastric inhibitory polypeptide receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Li, W.Z, Zhou, Q.T, Cong, Z.T, Yuan, Q.N, Li, W.X, Zhao, F.H, Xu, H.E, Zhao, L.H, Yang, D.H, Wang, M.W. | | Deposit date: | 2024-03-29 | | Release date: | 2024-09-18 | | Last modified: | 2024-09-25 | | Method: | ELECTRON MICROSCOPY (3.26 Å) | | Cite: | Structural insights into the triple agonism at GLP-1R, GIPR and GCGR manifested by retatrutide.
Cell Discov, 10, 2024
|
|
6LR6
 
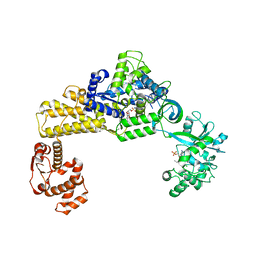 | | The crystal structure of human cytoplasmic LRS | | Descriptor: | 4-Chloro-3-aminomethyl-7-[ethoxy]-3H-benzo[C][1,2]oxaborol-1-ol modified adenosine, 5'-O-(L-leucylsulfamoyl)adenosine, Leucine--tRNA ligase, ... | | Authors: | Liu, R.J, Long, T, Li, H, Li, J, Zhao, J.H, Lin, J.Z, Palencia, A, Wang, M.Z, Cusack, S, Wang, E.D. | | Deposit date: | 2020-01-15 | | Release date: | 2020-03-25 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.009 Å) | | Cite: | Molecular basis of the multifaceted functions of human leucyl-tRNA synthetase in protein synthesis and beyond.
Nucleic Acids Res., 48, 2020
|
|
2QDY
 
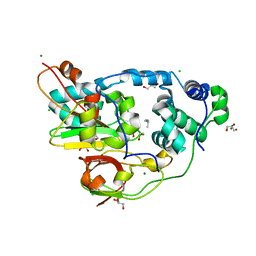 | | Crystal Structure of Fe-type NHase from Rhodococcus erythropolis AJ270 | | Descriptor: | 2-METHYLPROPAN-1-AMINE, CHLORIDE ION, FE (III) ION, ... | | Authors: | Song, L, Shi, J, Xue, Z, Wang, M.-X, Qian, S. | | Deposit date: | 2007-06-22 | | Release date: | 2008-05-13 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | High resolution X-ray molecular structure of the nitrile hydratase from Rhodococcus erythropolis AJ270 reveals posttranslational oxidation of two cysteines into sulfinic acids and a novel biocatalytic nitrile hydration mechanism
Biochem.Biophys.Res.Commun., 362, 2007
|
|
6LW5
 
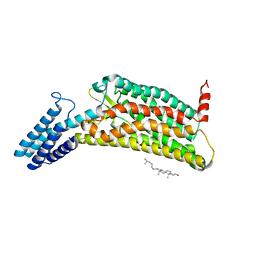 | | Crystal structure of the human formyl peptide receptor 2 in complex with WKYMVm | | Descriptor: | CHOLESTEROL, Soluble cytochrome b562,N-formyl peptide receptor 2, TRP-LYS-TYR-MET-VAL-QXV | | Authors: | Chen, T, Zong, X, Zhang, H, Wang, M, Zhao, Q, Wu, B. | | Deposit date: | 2020-02-07 | | Release date: | 2020-03-25 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural basis of ligand binding modes at the human formyl peptide receptor 2.
Nat Commun, 11, 2020
|
|
8JLV
 
 | |
6G89
 
 | | Thaumatin solved by Native SAD from a dataset collected in 0.6 second with JUNGFRAU detector | | Descriptor: | L(+)-TARTARIC ACID, Thaumatin I | | Authors: | Leonarski, F, Olieric, V, Vera, L, Redford, S, Wang, M. | | Deposit date: | 2018-04-08 | | Release date: | 2018-08-01 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.359 Å) | | Cite: | Fast and accurate data collection for macromolecular crystallography using the JUNGFRAU detector.
Nat. Methods, 15, 2018
|
|
6G8B
 
 | | E. coli Aminopeptidase N solved by Native SAD from a dataset collected in 60 second with JUNGFRAU detector | | Descriptor: | Aminopeptidase N, DIMETHYL SULFOXIDE, SODIUM ION, ... | | Authors: | Leonarski, F, Olieric, V, Redford, S, Wang, M. | | Deposit date: | 2018-04-08 | | Release date: | 2018-08-01 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.374 Å) | | Cite: | Fast and accurate data collection for macromolecular crystallography using the JUNGFRAU detector.
Nat. Methods, 15, 2018
|
|
4ZWJ
 
 | | Crystal structure of rhodopsin bound to arrestin by femtosecond X-ray laser | | Descriptor: | Chimera protein of human Rhodopsin, mouse S-arrestin, and T4 Endolysin | | Authors: | Kang, Y, Zhou, X.E, Gao, X, He, Y, Liu, W, Ishchenko, A, Barty, A, White, T.A, Yefanov, O, Han, G.W, Xu, Q, de Waal, P.W, Ke, J, Tan, M.H.E, Zhang, C, Moeller, A, West, G.M, Pascal, B, Eps, N.V, Caro, L.N, Vishnivetskiy, S.A, Lee, R.J, Suino-Powell, K.M, Gu, X, Pal, K, Ma, J, Zhi, X, Boutet, S, Williams, G.J, Messerschmidt, M, Gati, C, Zatsepin, N.A, Wang, D, James, D, Basu, S, Roy-Chowdhury, S, Conrad, C, Coe, J, Liu, H, Lisova, S, Kupitz, C, Grotjohann, I, Fromme, R, Jiang, Y, Tan, M, Yang, H, Li, J, Wang, M, Zheng, Z, Li, D, Howe, N, Zhao, Y, Standfuss, J, Diederichs, K, Dong, Y, Potter, C.S, Carragher, B, Caffrey, M, Jiang, H, Chapman, H.N, Spence, J.C.H, Fromme, P, Weierstall, U, Ernst, O.P, Katritch, V, Gurevich, V.V, Griffin, P.R, Hubbell, W.L, Stevens, R.C, Cherezov, V, Melcher, K, Xu, H.E, GPCR Network (GPCR) | | Deposit date: | 2015-05-19 | | Release date: | 2015-07-29 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.302 Å) | | Cite: | Crystal structure of rhodopsin bound to arrestin by femtosecond X-ray laser.
Nature, 523, 2015
|
|
6PB0
 
 | | Cryo-EM structure of Urocortin 1-bound Corticotropin-releasing factor 1 receptor in complex with Gs protein and Nb35 | | Descriptor: | CHOLESTEROL, Corticotropin-releasing factor receptor 1, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Ma, S, Shen, Q, Zhao, L.-H, Mao, C, Zhou, X.E, Shen, D.-D, de Waal, P.W, Bi, P, Li, C, Jiang, Y, Wang, M.-W, Sexton, P.M, Wootten, D, Melcher, K, Zhang, Y, Xu, H.E. | | Deposit date: | 2019-06-12 | | Release date: | 2020-02-12 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Molecular Basis for Hormone Recognition and Activation of Corticotropin-Releasing Factor Receptors.
Mol.Cell, 77, 2020
|
|
8IWF
 
 | | Aspergillus niger Rha-2 and pNPR | | Descriptor: | (2S,3R,4R,5R,6S)-2-methyl-6-[4-[oxidanyl(oxidanylidene)-$l^4-azanyl]phenoxy]oxane-3,4,5-triol, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Li, L.J, Li, L, Wang, M.H, Jiang, Z.D, Zhu, Y.B, Jin, T.C, Ni, H. | | Deposit date: | 2023-03-29 | | Release date: | 2024-04-03 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.86 Å) | | Cite: | Crystal structure and catalytic function of alpha-L-rhamnosidase from Aspergillus niger
To Be Published
|
|
8IWD
 
 | | Aspergillus niger Rha-2 | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Li, L.J, Li, L, Wang, M.H, Jiang, Z.D, Zhu, Y.B, Jin, T.C, Ni, H. | | Deposit date: | 2023-03-29 | | Release date: | 2024-04-03 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | Crystal structure and catalytic function of alpha-L-rhamnosidase from Aspergillus niger
To Be Published
|
|
6PB1
 
 | | Cryo-EM structure of Urocortin 1-bound Corticotropin-releasing factor 2 receptor in complex with Gs protein and Nb35 | | Descriptor: | CHOLESTEROL, Corticotropin-releasing factor receptor 2, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Ma, S, Shen, Q, Zhao, L.-H, Mao, C, Zhou, X.E, Shen, D.-D, de Waal, P.W, Bi, P, Li, C, Jiang, Y, Wang, M.-W, Sexton, P.M, Wootten, D, Melcher, K, Zhang, Y, Xu, H.E. | | Deposit date: | 2019-06-12 | | Release date: | 2020-02-12 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Molecular Basis for Hormone Recognition and Activation of Corticotropin-Releasing Factor Receptors.
Mol.Cell, 77, 2020
|
|
4XNJ
 
 | | X-ray structure of PepTst2 | | Descriptor: | (2S)-2,3-DIHYDROXYPROPYL(7Z)-PENTADEC-7-ENOATE, Di-or tripeptide:H+ symporter, PHOSPHATE ION | | Authors: | Huang, C.Y, Olieric, V, Diederichs, K, Wang, M, Caffrey, M. | | Deposit date: | 2015-01-15 | | Release date: | 2015-06-03 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | In meso in situ serial X-ray crystallography of soluble and membrane proteins.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4XNL
 
 | | X-ray structure of AlgE2 | | Descriptor: | (2R)-2,3-DIHYDROXYPROPYL(7Z)-PENTADEC-7-ENOATE, (2S)-2,3-DIHYDROXYPROPYL(7Z)-PENTADEC-7-ENOATE, 3,6,9,12,15,18,21,24-OCTAOXAHEXACOSAN-1-OL, ... | | Authors: | Ma, P, Huang, C.Y, Olieric, V, Diederichs, K, Wang, M, Caffrey, M. | | Deposit date: | 2015-01-15 | | Release date: | 2015-06-03 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | In meso in situ serial X-ray crystallography of soluble and membrane proteins.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4XNI
 
 | | X-ray structure of PepTst1 | | Descriptor: | (2S)-2,3-DIHYDROXYPROPYL(7Z)-PENTADEC-7-ENOATE, Di-or tripeptide:H+ symporter, PHOSPHATE ION | | Authors: | Huang, C.Y, Olieric, V, Diederichs, K, Wang, M, Caffrey, M. | | Deposit date: | 2015-01-15 | | Release date: | 2015-06-03 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | In meso in situ serial X-ray crystallography of soluble and membrane proteins.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
8AQ2
 
 | | In meso structure of the membrane integral lipoprotein N-acyltransferase Lnt from P. aeruginosa covalently linked with TITC | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Apolipoprotein N-acyltransferase, CITRATE ANION, ... | | Authors: | Huang, C.-Y, Weichert, D, Boland, C, Smithers, L, Olieric, V, Wang, M, Caffrey, M. | | Deposit date: | 2022-08-11 | | Release date: | 2023-07-12 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure snapshots reveal the mechanism of a bacterial membrane lipoprotein N -acyltransferase.
Sci Adv, 9, 2023
|
|
8AQ3
 
 | | In surfo structure of the membrane integral lipoprotein N-acyltransferase Lnt from E. coli in complex with PE | | Descriptor: | 1-(2-METHOXY-ETHOXY)-2-{2-[2-(2-METHOXY-ETHOXY]-ETHOXY}-ETHANE, 1-METHOXY-2-[2-(2-METHOXY-ETHOXY]-ETHANE, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ... | | Authors: | Huang, C.-Y, Weichert, D, Boland, C, Smithers, L, Olieric, V, Wang, M, Caffrey, M. | | Deposit date: | 2022-08-11 | | Release date: | 2023-07-12 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.395 Å) | | Cite: | Structure snapshots reveal the mechanism of a bacterial membrane lipoprotein N -acyltransferase.
Sci Adv, 9, 2023
|
|
8AQ4
 
 | | In surfo structure of the membrane integral lipoprotein N-acyltransferase Lnt from E. coli in complex with TITC and lyso-PE | | Descriptor: | 1-(2-METHOXY-ETHOXY)-2-{2-[2-(2-METHOXY-ETHOXY]-ETHOXY}-ETHANE, 1-METHOXY-2-[2-(2-METHOXY-ETHOXY]-ETHANE, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ... | | Authors: | Huang, C.-Y, Weichert, D, Boland, C, Smithers, L, Olieric, V, Wang, M, Caffrey, M. | | Deposit date: | 2022-08-11 | | Release date: | 2023-07-12 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.62 Å) | | Cite: | Structure snapshots reveal the mechanism of a bacterial membrane lipoprotein N -acyltransferase.
Sci Adv, 9, 2023
|
|
