3L7O
 
 | |
4M48
 
 | | X-ray structure of dopamine transporter elucidates antidepressant mechanism | | Descriptor: | 9D5 antibody, heavy chain, light chain, ... | | Authors: | Gouaux, E, Penmatsa, A, Wang, K. | | Deposit date: | 2013-08-06 | | Release date: | 2013-09-18 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.955 Å) | | Cite: | X-ray structure of dopamine transporter elucidates antidepressant mechanism.
Nature, 503, 2013
|
|
7XB2
 
 | |
7WL3
 
 | | CVB5 expended empty particle | | Descriptor: | Capsid protein, Genome polyprotein | | Authors: | Yang, P, Wang, K. | | Deposit date: | 2022-01-12 | | Release date: | 2022-03-30 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (2.95 Å) | | Cite: | Atomic Structures of Coxsackievirus B5 Provide Key Information on Viral Evolution and Survival.
J.Virol., 96, 2022
|
|
7F3Q
 
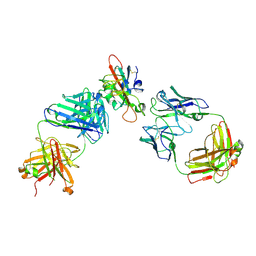 | | SARS-CoV-2 RBD in complex with A5-10 Fab and A34-2 Fab | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of A34-2 Fab, ... | | Authors: | Dou, Y, Wang, X, Liu, P, Lu, B, Wang, K. | | Deposit date: | 2021-06-16 | | Release date: | 2022-06-22 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Development of neutralizing antibodies against SARS-CoV-2, using a high-throughput single-B-cell cloning method.
Antib Ther, 6, 2023
|
|
8WA4
 
 | |
5XHX
 
 | |
3RSN
 
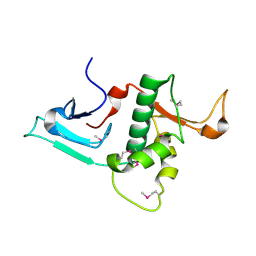 | | Crystal Structure of the N-terminal region of Human Ash2L | | Descriptor: | Set1/Ash2 histone methyltransferase complex subunit ASH2, ZINC ION | | Authors: | Chen, Y, Wan, B, Wang, K.C, Cao, F, Yang, Y, Protacio, A, Dou, Y, Chang, H.Y, Lei, M. | | Deposit date: | 2011-05-02 | | Release date: | 2011-06-22 | | Last modified: | 2011-12-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of the N-terminal region of human Ash2L shows a winged-helix motif involved in DNA binding.
Embo Rep., 12, 2011
|
|
1C35
 
 | | SOLUTION STRUCTURE OF A QUADRUPLEX FORMING DNA AND ITS INTERMIDIATE | | Descriptor: | DNA (5'-D(*GP*GP*TP*TP*GP*GP*TP*GP*TP*GP*GP*TP*TP*GP*G)-3'), POTASSIUM ION | | Authors: | Bolton, P.H, Marathias, V.M, Wang, K. | | Deposit date: | 1999-07-25 | | Release date: | 1999-08-18 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structures of the potassium-saturated, 2:1, and intermediate, 1:1, forms of a quadruplex DNA.
Nucleic Acids Res., 28, 2000
|
|
1C38
 
 | | SOLUTION STRUCTURE OF A QUADRUPLEX FORMING DNA AND ITS INTERMEDIATE | | Descriptor: | DNA (5'-D(*GP*GP*TP*TP*GP*GP*TP*GP*TP*GP*GP*TP*TP*GP*G)-3'), POTASSIUM ION | | Authors: | Bolton, P.H, Marathias, V.M, Wang, K. | | Deposit date: | 1999-07-25 | | Release date: | 1999-08-25 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structures of the potassium-saturated, 2:1, and intermediate, 1:1, forms of a quadruplex DNA.
Nucleic Acids Res., 28, 2000
|
|
4OCO
 
 | | N-acetylhexosamine 1-phosphate kinase in complex with GlcNAc-1-phosphate | | Descriptor: | 2-acetamido-2-deoxy-1-O-phosphono-alpha-D-glucopyranose, N-acetylhexosamine 1-phosphate kinase | | Authors: | Li, T.L, Wang, K.C, Lyu, S.Y, Liu, Y.C, Chang, C.Y, Wu, C.J. | | Deposit date: | 2014-01-09 | | Release date: | 2014-05-14 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Insights into the binding specificity and catalytic mechanism of N-acetylhexosamine 1-phosphate kinases through multiple reaction complexes.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4OCJ
 
 | | N-acetylhexosamine 1-phosphate kinase in complex with GlcNAc | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose, N-acetylhexosamine 1-phosphate kinase, SODIUM ION | | Authors: | Li, T.L, Wang, K.C, Lyu, S.Y, Liu, Y.C, Chang, C.Y, Wu, C.J. | | Deposit date: | 2014-01-09 | | Release date: | 2014-05-14 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.571 Å) | | Cite: | Insights into the binding specificity and catalytic mechanism of N-acetylhexosamine 1-phosphate kinases through multiple reaction complexes.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4OCU
 
 | | N-acetylhexosamine 1-phosphate kinase_ATCC15697 in complex with GlcNAc | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose, N-acetylhexosamine 1-phosphate kinase, SULFATE ION | | Authors: | Li, T.L, Wang, K.C, Lyu, S.Y, Liu, Y.C, Chang, C.Y, Wu, C.J. | | Deposit date: | 2014-01-09 | | Release date: | 2014-05-14 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.904 Å) | | Cite: | Insights into the binding specificity and catalytic mechanism of N-acetylhexosamine 1-phosphate kinases through multiple reaction complexes.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
2L01
 
 | | Solution NMR Structure of protein BVU3908 from Bacteroides vulgatus, Northeast Structural Genomics Consortium Target BvR153 | | Descriptor: | Uncharacterized protein | | Authors: | Eletsky, A, Lee, C, Wang, K, Ciccosanti, T.B, Hamilton, R, Acton, J.B, Xiao, G.B, Everett, J.K, Prestegard, J.H, Montelione, G.T, Szyperski, T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2010-06-29 | | Release date: | 2010-08-25 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution NMR Structure of protein BVU3908 from Bacteroides vulgatus
To be Published
|
|
8HHT
 
 | | Crystal structure of the SARS-CoV-2 main protease in complex with Hit-1 | | Descriptor: | 3C-like proteinase nsp5, DIMETHYL SULFOXIDE, ~{N}-[(2~{R},3~{S})-3-oxidanyl-4-oxidanylidene-1-phenyl-4-(1,3-thiazol-2-ylmethylamino)butan-2-yl]benzamide | | Authors: | Zeng, R, Xie, L.W, Huang, C, Wang, K, Liu, Y.Z, Yang, S.Y, Lei, J. | | Deposit date: | 2022-11-17 | | Release date: | 2023-03-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | A new generation M pro inhibitor with potent activity against SARS-CoV-2 Omicron variants.
Signal Transduct Target Ther, 8, 2023
|
|
8HHU
 
 | | Crystal structure of the SARS-CoV-2 main protease in complex with SY110 | | Descriptor: | (1~{R})-3,3-bis(fluoranyl)-~{N}-[(2~{R})-3-methoxy-1-oxidanylidene-1-[[(2~{R},3~{S})-3-oxidanyl-4-oxidanylidene-1-phenyl-4-(1,3-thiazol-2-ylmethylamino)butan-2-yl]amino]propan-2-yl]cyclohexane-1-carboxamide, 3C-like proteinase nsp5 | | Authors: | Zeng, R, Xie, L.W, Huang, C, Wang, K, Liu, Y.Z, Yang, S.Y, Lei, J. | | Deposit date: | 2022-11-17 | | Release date: | 2023-03-29 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.258 Å) | | Cite: | A new generation M pro inhibitor with potent activity against SARS-CoV-2 Omicron variants.
Signal Transduct Target Ther, 8, 2023
|
|
4OCK
 
 | | N-acetylhexosamine 1-phosphate kinase in complex with GlcNAc and AMPPNP | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose, N-acetylhexosamine 1-phosphate kinase, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER | | Authors: | Li, T.L, Wang, K.C, Lyu, S.Y, Liu, Y.C, Chang, C.Y, Wu, C.J. | | Deposit date: | 2014-01-09 | | Release date: | 2014-05-14 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Insights into the binding specificity and catalytic mechanism of N-acetylhexosamine 1-phosphate kinases through multiple reaction complexes.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4OCP
 
 | | N-acetylhexosamine 1-phosphate kinase in complex with GlcNAc-1-phosphate and ADP | | Descriptor: | 2-acetamido-2-deoxy-1-O-phosphono-alpha-D-glucopyranose, ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Li, T.L, Wang, K.C, Lyu, S.Y, Liu, Y.C, Chang, C.Y, Wu, C.J. | | Deposit date: | 2014-01-09 | | Release date: | 2014-05-14 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.938 Å) | | Cite: | Insights into the binding specificity and catalytic mechanism of N-acetylhexosamine 1-phosphate kinases through multiple reaction complexes.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4OCV
 
 | | N-acetylhexosamine 1-phosphate kinase_ATCC15697 in complex with GlcNAc and AMPPNP | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose, N-acetylhexosamine 1-phosphate kinase, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Li, T.L, Wang, K.C, Lyu, S.Y, Liu, Y.C, Chang, C.Y, Wu, C.J. | | Deposit date: | 2014-01-09 | | Release date: | 2014-05-14 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.472 Å) | | Cite: | Insights into the binding specificity and catalytic mechanism of N-acetylhexosamine 1-phosphate kinases through multiple reaction complexes.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4OCQ
 
 | | N-acetylhexosamine 1-phosphate kinase in complex with GalNAc | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-galactopyranose, N-acetylhexosamine 1-phosphate kinase | | Authors: | Li, T.L, Wang, K.C, Lyu, S.Y, Liu, Y.C, Chang, C.Y, Wu, C.J. | | Deposit date: | 2014-01-09 | | Release date: | 2014-05-14 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.878 Å) | | Cite: | Insights into the binding specificity and catalytic mechanism of N-acetylhexosamine 1-phosphate kinases through multiple reaction complexes.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
7C9Z
 
 | | Coxsackievirus B1 F-particle | | Descriptor: | MYRISTIC ACID, PALMITIC ACID, VP1, ... | | Authors: | Feng, R, Wang, K, Rao, Z, Wang, X. | | Deposit date: | 2020-06-08 | | Release date: | 2020-08-12 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structures of Echovirus 30 in complex with its receptors inform a rational prediction for enterovirus receptor usage.
Nat Commun, 11, 2020
|
|
3VZX
 
 | | Crystal structure of PcrB from bacillus subtilis subap. subtilis str. 168 | | Descriptor: | CHLORIDE ION, Heptaprenylglyceryl phosphate synthase, MAGNESIUM ION | | Authors: | Ren, F, Feng, X, Ko, T.P, Huang, C.H, Hu, Y, Chan, H.C, Liu, Y.L, Wang, K, Chen, C.C, Pang, X, He, M, Li, Y, Oldfield, E, Guo, R.T. | | Deposit date: | 2012-10-17 | | Release date: | 2012-12-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Insights into TIM-barrel prenyl transferase mechanisms: crystal structures of PcrB from Bacillus subtilis and Staphylococcus aureus
Chembiochem, 14, 2013
|
|
3VZY
 
 | | Crystal structure of PcrB complexed with G1P from bacillus subtilis subap. subtilis str. 168 | | Descriptor: | CHLORIDE ION, Heptaprenylglyceryl phosphate synthase, MAGNESIUM ION, ... | | Authors: | Ren, F, Feng, X, Ko, T.P, Huang, C.H, Hu, Y, Chan, H.C, Liu, Y.L, Wang, K, Chen, C.C, Pang, X, He, M, Li, Y, Oldfield, E, Guo, R.T. | | Deposit date: | 2012-10-17 | | Release date: | 2012-12-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Insights into TIM-barrel prenyl transferase mechanisms: crystal structures of PcrB from Bacillus subtilis and Staphylococcus aureus
Chembiochem, 14, 2013
|
|
3V97
 
 | |
3W01
 
 | | Crystal structure of PcrB complexed with PEG from Staphylococcus aureus subsp. aureus Mu3 | | Descriptor: | Heptaprenylglyceryl phosphate synthase, TRIETHYLENE GLYCOL | | Authors: | Ren, F, Feng, X, Ko, T.P, Huang, C.H, Hu, Y, Chan, H.C, Liu, Y.L, Wang, K, Chen, C.C, Pang, X, He, M, Li, Y, Oldfield, E, Guo, R.T. | | Deposit date: | 2012-10-17 | | Release date: | 2012-12-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Insights into TIM-barrel prenyl transferase mechanisms: crystal structures of PcrB from Bacillus subtilis and Staphylococcus aureus
Chembiochem, 14, 2013
|
|
