5XLN
 
 | |
4V24
 
 | | Sphingosine kinase 1 in complex with PF-543 | | Descriptor: | ACETATE ION, SPHINGOSINE KINASE 1, {(2R)-1-[4-({3-METHYL-5-[(PHENYLSULFONYL)METHYL]PHENOXY}METHYL)BENZYL]PYRROLIDIN-2-YL}METHANOL | | Authors: | Elkins, J.M, Wang, J, Sorrell, F, Tallant, C, Wang, D, Shrestha, L, Bountra, C, von Delft, F, Knapp, S, Edwards, A. | | Deposit date: | 2014-10-05 | | Release date: | 2014-10-15 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of Sphingosine Kinase 1 with Pf-543.
Acs Med.Chem.Lett., 5, 2014
|
|
2PQE
 
 | | Solution structure of proline-free mutant of staphylococcal nuclease | | Descriptor: | Thermonuclease | | Authors: | Shan, L, Tong, Y, Xie, T, Wang, M, Wang, J. | | Deposit date: | 2007-05-01 | | Release date: | 2007-06-19 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Restricted backbone conformational and motional flexibilities of loops containing peptidyl-proline bonds dominate the enzyme activity of staphylococcal nuclease.
Biochemistry, 46, 2007
|
|
3J9B
 
 | | Electron cryo-microscopy of an RNA polymerase | | Descriptor: | Polymerase, Polymerase basic protein 2, RNA (5'-R(*UP*UP*UP*UP*UP*A)-3'), ... | | Authors: | Chang, S.H, Sun, D.P, Liang, H.H, Wang, J, Li, J, Guo, L, Wang, X.L, Guan, C.C, Boruah, B.M, Yuan, L.M, Feng, F, Yang, M.R, Wojdyla, J, Wang, J.W, Wang, M.T, Wang, H.W, Liu, Y.F. | | Deposit date: | 2014-12-16 | | Release date: | 2015-02-18 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Cryo-EM Structure of Influenza Virus RNA Polymerase Complex at 4.3 angstrom Resolution.
Mol.Cell, 2015
|
|
6Z71
 
 | | Structure of the MATE family multidrug resistance transporter Aq_128 from Aquifex aeolicus in the outward-facing state | | Descriptor: | Aq128 | | Authors: | Zhao, J, Safarian, S, Thielmann, Y, Xie, H, Wang, J, Michel, H. | | Deposit date: | 2020-05-29 | | Release date: | 2021-12-01 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | The structure of the Aquifex aeolicus MATE family multidrug resistance transporter and sequence comparisons suggest the existence of a new subfamily.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
6Z70
 
 | | Structure of the MATE family multidrug resistance transporter Aq_128 from Aquifex aeolicus in the outward-facing state | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Aq128 | | Authors: | Zhao, J, Safarian, S, Thielmann, Y, Xie, H, Wang, J, Michel, H. | | Deposit date: | 2020-05-29 | | Release date: | 2021-12-01 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The structure of the Aquifex aeolicus MATE family multidrug resistance transporter and sequence comparisons suggest the existence of a new subfamily.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
3ZON
 
 | | Human TYK2 pseudokinase domain bound to a kinase inhibitor | | Descriptor: | 5-PHENYL-2-UREIDOTHIOPHENE-3-CARBOXAMIDE, NON-RECEPTOR TYROSINE-PROTEIN KINASE TYK2 | | Authors: | Elkins, J.M, Wang, J, Krojer, T, Savitsky, P, Chalk, R, Daga, N, Salah, E, Berridge, G, Picaud, S, von Delft, F, Bountra, C, Edwards, A, Knapp, S. | | Deposit date: | 2013-02-22 | | Release date: | 2013-04-10 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Human Tyk2 Pseudokinase Domain Bound to a Kinase Inhibitor
To be Published
|
|
1QRZ
 
 | | CATALYTIC DOMAIN OF PLASMINOGEN | | Descriptor: | PLASMINOGEN | | Authors: | Peisach, E, Wang, J, de los Santos, T, Reich, E, Ringe, D. | | Deposit date: | 1999-06-16 | | Release date: | 1999-10-14 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of the proenzyme domain of plasminogen.
Biochemistry, 38, 1999
|
|
2Q06
 
 | | Crystal structure of Influenza A Virus H5N1 Nucleoprotein | | Descriptor: | Nucleoprotein | | Authors: | Ng, A.K.L, Zhang, H, Tan, K, Wang, J, Shaw, P.C. | | Deposit date: | 2007-05-21 | | Release date: | 2008-05-27 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structure of the influenza virus A H5N1 nucleoprotein: implications for RNA binding, oligomerization, and vaccine design.
Faseb J., 22, 2008
|
|
1R5H
 
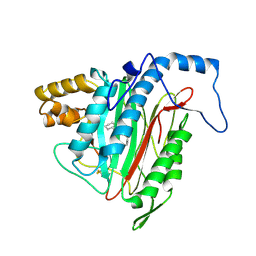 | | Crystal Structure of MetAP2 complexed with A320282 | | Descriptor: | MANGANESE (II) ION, Methionine aminopeptidase 2, N'-(2S,3R)-3-AMINO-4-CYCLOHEXYL-2-HYDROXY-BUTANO-N-(4-METHYLPHENYL)HYDRAZIDE | | Authors: | Sheppard, G.S, Wang, J, Kawai, M, BaMaung, N.Y, Craig, R.A, Erickson, S.A, Lynch, L, Patel, J, Yang, F, Searle, X.B, Lou, P, Park, C, Kim, K.H, Henkin, J, Lesniewski, R. | | Deposit date: | 2003-10-10 | | Release date: | 2004-10-12 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | 3-Amino-2-hydroxyamides and related compounds as inhibitors of methionine aminopeptidase-2.
Bioorg.Med.Chem.Lett., 14, 2004
|
|
1R58
 
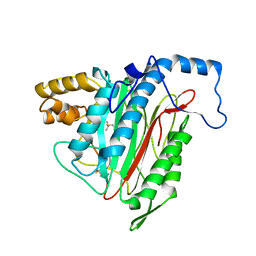 | | Crystal Structure of MetAP2 complexed with A357300 | | Descriptor: | MANGANESE (II) ION, Methionine aminopeptidase 2, N'-((2S,3R)-3-AMINO-2-HYDROXY-5-(ISOPROPYLSULFANYL)PENTANOYL)-N-3-CHLOROBENZOYL HYDRAZIDE | | Authors: | Sheppard, G.S, Wang, J, Kawai, M, BaMaung, N.Y, Craig, R.A, Ericken, S.A, Lynch, L, Patel, J, Yang, F, Searle, X.B, Lou, P, Park, C, Kim, K.H, Henkin, J, Lesniewski, R. | | Deposit date: | 2003-10-09 | | Release date: | 2004-10-12 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | 3-Amino-2-hydroxyamides and related compounds as inhibitors of methionine aminopeptidase-2.
Bioorg.Med.Chem.Lett., 14, 2004
|
|
3KCU
 
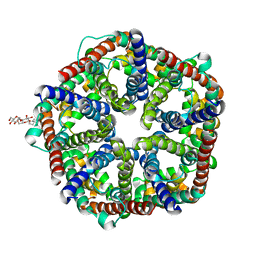 | | Structure of formate channel | | Descriptor: | 2-(6-(2-CYCLOHEXYLETHOXY)-TETRAHYDRO-4,5-DIHYDROXY-2(HYDROXYMETHYL)-2H-PYRAN-3-YLOXY)-TETRAHYDRO-6(HYDROXYMETHYL)-2H-PY RAN-3,4,5-TRIOL, Probable formate transporter 1 | | Authors: | Wang, Y, Huang, Y, Wang, J, Yan, N, Shi, Y. | | Deposit date: | 2009-10-22 | | Release date: | 2009-12-01 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.243 Å) | | Cite: | Structure of the formate transporter FocA reveals a pentameric aquaporin-like channel
Nature, 462, 2009
|
|
3KCV
 
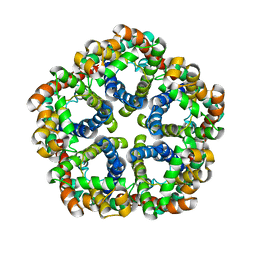 | | Structure of formate channel | | Descriptor: | Probable formate transporter 1 | | Authors: | Wang, Y, Huang, Y, Wang, J, Yan, N, Shi, Y. | | Deposit date: | 2009-10-22 | | Release date: | 2009-12-01 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.198 Å) | | Cite: | Structure of the formate transporter FocA reveals a pentameric aquaporin-like channel
Nature, 462, 2009
|
|
9IUZ
 
 | | Constitutively active mutant(Y276H) of Arabidopsis phytochrome B(phyB) in complex with phytochrome-interacting factor 6(PIF6) | | Descriptor: | 3-[5-[[(3~{R},4~{R})-3-ethyl-4-methyl-5-oxidanylidene-3,4-dihydropyrrol-2-yl]methyl]-2-[[5-[(4-ethyl-3-methyl-5-oxidanylidene-pyrrol-2-yl)methyl]-3-(3-hydroxy-3-oxopropyl)-4-methyl-1~{H}-pyrrol-2-yl]methyl]-4-methyl-1~{H}-pyrrol-3-yl]propanoic acid, Phytochrome B, Phytochrome-interacting factor 6 | | Authors: | Wang, Z, Wang, W, Zhao, D, Song, Y, Xu, B, Zhao, J, Wang, J. | | Deposit date: | 2024-07-22 | | Release date: | 2024-10-02 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.19 Å) | | Cite: | Light-induced remodeling of phytochrome B enables signal transduction by phytochrome-interacting factor.
Cell, 2024
|
|
8YB4
 
 | | Pfr conformer of Arabidopsis thaliana phytochrome B in complex with phytochrome-interacting factor 6 | | Descriptor: | 3-[5-[[(3~{R},4~{R})-3-ethyl-4-methyl-5-oxidanylidene-3,4-dihydropyrrol-2-yl]methyl]-2-[[5-[(4-ethyl-3-methyl-5-oxidanylidene-pyrrol-2-yl)methyl]-3-(3-hydroxy-3-oxopropyl)-4-methyl-1~{H}-pyrrol-2-yl]methyl]-4-methyl-1~{H}-pyrrol-3-yl]propanoic acid, phytochrome B, phytochrome-interacting factor 6 | | Authors: | Wang, Z, Wang, W, Zhao, D, Song, Y, Xu, B, Zhao, J, Wang, J. | | Deposit date: | 2024-02-11 | | Release date: | 2024-10-02 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Light-induced remodeling of phytochrome B enables signal transduction by phytochrome-interacting factor.
Cell, 2024
|
|
7LU6
 
 | | Crystal structure of the mouse Kirrel3 D1 homodimer | | Descriptor: | Kin of IRRE-like protein 3, SODIUM ION | | Authors: | Roman, C.A, Pak, J.S, Wang, J, Ozkan, E. | | Deposit date: | 2021-02-21 | | Release date: | 2021-11-24 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Molecular and structural basis of olfactory sensory neuron axon coalescence by Kirrel receptors.
Cell Rep, 37, 2021
|
|
7LTW
 
 | | Crystal structure of the mouse Kirrel2 D1 homodimer | | Descriptor: | Kin of IRRE-like protein 2, SODIUM ION | | Authors: | Roman, C.A, Pak, J.S, Wang, J, Ozkan, E. | | Deposit date: | 2021-02-20 | | Release date: | 2021-11-24 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Molecular and structural basis of olfactory sensory neuron axon coalescence by Kirrel receptors.
Cell Rep, 37, 2021
|
|
7LW2
 
 | |
6E5V
 
 | | human mGlu8 receptor amino terminal domain in complex with (S)-3,4-Dicarboxyphenylglycine (DCPG) | | Descriptor: | 4-[(S)-amino(carboxy)methyl]benzene-1,2-dicarboxylic acid, CHLORIDE ION, Metabotropic glutamate receptor 8 | | Authors: | Chen, Q, Ho, J.D, Ashok, S, Vargas, M.C, Wang, J, Atwell, S, Bures, M, Schkeryantz, J.M, Monn, J.A, Hao, J. | | Deposit date: | 2018-07-23 | | Release date: | 2018-11-07 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Structural Basis for ( S)-3,4-Dicarboxyphenylglycine (DCPG) As a Potent and Subtype Selective Agonist of the mGlu8Receptor.
J. Med. Chem., 61, 2018
|
|
4N76
 
 | | Structure of Thermus thermophilus Argonaute bound to guide DNA and cleaved target DNA with Mn2+ | | Descriptor: | 5'-D(P*TP*AP*CP*TP*AP*CP*CP*TP*CP*G)-3', 5'-D(P*TP*GP*AP*GP*GP*TP*AP*GP*TP*AP*GP*GP*TP*TP*GP*TP*AP*TP*AP*GP*T)-3', Argonaute, ... | | Authors: | Sheng, G, Zhao, H, Wang, J, Rao, Y, Wang, Y. | | Deposit date: | 2013-10-15 | | Release date: | 2014-01-15 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.89 Å) | | Cite: | Structure-based cleavage mechanism of Thermus thermophilus Argonaute DNA guide strand-mediated DNA target cleavage.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
6E59
 
 | | Crystal structure of the human NK1 tachykinin receptor | | Descriptor: | 1-(4-{[(2R,3S)-2-{(1R)-1-[3,5-bis(trifluoromethyl)phenyl]ethoxy}-3-(4-fluorophenyl)morpholin-4-yl]methyl}-1H-1,2,3-triazol-5-yl)-N,N-dimethylmethanamine, Substance-P receptor, GlgA glycogen synthase, ... | | Authors: | Yin, J, Clark, L, Chapman, K, Shao, Z, Borek, D, Xu, Q, Wang, J, Rosenbaum, D.M. | | Deposit date: | 2018-07-19 | | Release date: | 2018-12-12 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Crystal structure of the human NK1tachykinin receptor.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
2A5J
 
 | | Crystal Structure of Human RAB2B | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Ras-related protein Rab-2B | | Authors: | Dong, A, Wang, J, Shen, Y, Arrowsmith, C.H, Edwards, A.M, Sundstrom, M, Bochkarev, A, Park, H.W, Structural Genomics Consortium (SGC) | | Deposit date: | 2005-06-30 | | Release date: | 2005-07-19 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.501 Å) | | Cite: | Crystal structure of human RAB2B
To be Published
|
|
3KHF
 
 | | The crystal structure of the PDZ domain of human Microtubule Associated Serine/Threonine Kinase 3 (MAST3) | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Microtubule-associated serine/threonine-protein kinase 3 | | Authors: | Roos, A, Elkins, J, Savitsky, P, Wang, J, Ugochukwu, E, Murray, J, Bountra, C, Arrowsmith, C.H, Weigelt, J, Edwards, A, von Delft, F, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-10-30 | | Release date: | 2009-12-08 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | The crystal structure of the PDZ domain of human Microtubule Associated Serine/Threonine Kinase 3 (MAST3)
To be Published
|
|
6T7K
 
 | | Crystal Structure of Prolyl-tRNA synthetase (ProRS, Proline--tRNA ligase) from Plasmodium falciparum in complex with NCP-26 and L-Proline | | Descriptor: | 1,2-ETHANEDIOL, PROLINE, Proline--tRNA ligase, ... | | Authors: | Johansson, C, Wang, J, Tye, M, Payne, N.C, Mazitschek, R, Thompson, A, Arrowsmith, C.H, Bountra, C, Edwards, A, Oppermann, U.C.T. | | Deposit date: | 2019-10-22 | | Release date: | 2020-11-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Crystal Structure of Prolyl-tRNA synthetase (ProRS, Proline--tRNA ligase) from Plasmodium falciparum in complex with NCP-26 and L-Proline
To Be Published
|
|
5WTH
 
 | | Cryo-EM structure for Hepatitis A virus complexed with FAB | | Descriptor: | FAB Heavy Chain, FAB Light Chain, Polyprotein, ... | | Authors: | Wang, X, Zhu, L, Dang, M, Hu, Z, Gao, Q, Yuan, S, Sun, Y, Zhang, B, Ren, J, Walter, T.S, Wang, J, Fry, E.E, Stuart, D.I, Rao, Z. | | Deposit date: | 2016-12-12 | | Release date: | 2017-01-25 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Potent neutralization of hepatitis A virus reveals a receptor mimic mechanism and the receptor recognition site
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
