5VIY
 
 | |
4IQ8
 
 | | Crystal structure of glyceraldehyde-3-phosphate dehydrogenase 3 from Saccharomyces cerevisiae | | Descriptor: | Glyceraldehyde-3-phosphate dehydrogenase 3 | | Authors: | Wang, H, Liu, Q, Niu, L, Teng, M, Li, X. | | Deposit date: | 2013-01-11 | | Release date: | 2013-02-06 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Preliminary crystallographic analysis of glyceraldehyde-3-phosphate dehydrogenase 3 from Saccharomyces cerevisiae.
Acta Crystallogr.,Sect.F, 68, 2012
|
|
6KA1
 
 | | E.coli Malate dehydrogenase | | Descriptor: | Malate dehydrogenase | | Authors: | Wang, H, Wang, M, Sun, H. | | Deposit date: | 2019-06-19 | | Release date: | 2020-06-17 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.543 Å) | | Cite: | MDH is a major silver target in E. coli
To Be Published
|
|
5THR
 
 | | Cryo-EM structure of a BG505 Env-sCD4-17b-8ANC195 complex | | Descriptor: | 17b Fab VH domain, 17b Fab VL domain, 8ANC195 G52K5 VH domain, ... | | Authors: | Wang, H, Bjorkman, P.J. | | Deposit date: | 2016-09-30 | | Release date: | 2016-11-16 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (8.9 Å) | | Cite: | Cryo-EM structure of a CD4-bound open HIV-1 envelope trimer reveals structural rearrangements of the gp120 V1V2 loop.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5DGI
 
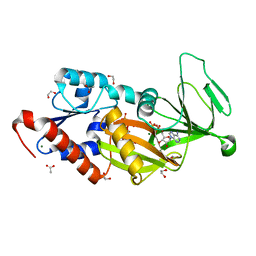 | | Crystal structure of the catalytic domain of human diphosphoinositol pentakisphosphate kinase 2 (PPIP5K2) in complex with ADP and 3,5-(PCP)2-IP4 | | Descriptor: | 1,2-ETHANEDIOL, 3,5-di-methylenebisphosphonate inositol tetrakisphosphate, ACETATE ION, ... | | Authors: | Wang, H, Shears, S.B. | | Deposit date: | 2015-08-27 | | Release date: | 2016-08-10 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Cellular Cations Control Conformational Switching of Inositol Pyrophosphate Analogues.
Chemistry, 22, 2016
|
|
5VJ6
 
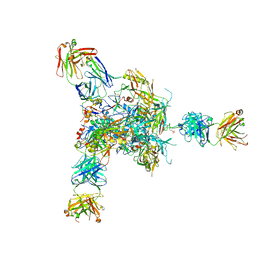 | |
4EGL
 
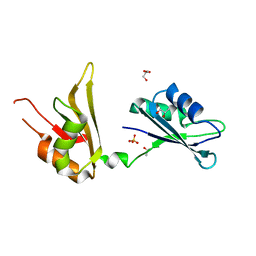 | | Crystal structure of two tandem RNA recognition motifs of Human antigen R | | Descriptor: | ELAV-like protein 1, GLYCEROL, SULFATE ION | | Authors: | Wang, H, Zeng, F, Liu, H, Teng, M, Li, X. | | Deposit date: | 2012-03-31 | | Release date: | 2012-05-30 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structure of two tandem RNA recognition motifs of Human antigen R
To be Published
|
|
5W2I
 
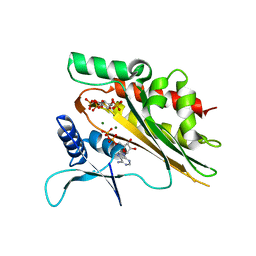 | | Crystal structure of the core catalytic domain of human inositol phosphate multikinase soaked with C4-analogue of PtdIns(4,5)P2 and ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, D-MYO-INOSITOL-1,4,5-TRIPHOSPHATE, Inositol polyphosphate multikinase,Inositol polyphosphate multikinase, ... | | Authors: | Wang, H, Shears, S.B. | | Deposit date: | 2017-06-06 | | Release date: | 2017-09-13 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural features of human inositol phosphate multikinase rationalize its inositol phosphate kinase and phosphoinositide 3-kinase activities.
J. Biol. Chem., 292, 2017
|
|
5DGH
 
 | |
6BYF
 
 | |
5Z3W
 
 | | Malate dehydrogenase binds silver at C113 | | Descriptor: | Malate dehydrogenase, SILVER ION | | Authors: | Wang, H, Wang, M, Sun, H. | | Deposit date: | 2018-01-09 | | Release date: | 2019-01-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Atomic differentiation of silver binding preference in protein targets: Escherichia coli malate dehydrogenase as a paradigm
Chem Sci, 2020
|
|
5W2G
 
 | |
5F18
 
 | | Structural basis of Ebola virus entry: viral glycoprotein bound to its endosomal receptor Niemann-Pick C1 | | Descriptor: | Niemann-Pick C1 protein | | Authors: | Wang, H, Shi, Y, Song, J, Qi, J, Lu, G, Yan, J, Gao, G.F. | | Deposit date: | 2015-11-30 | | Release date: | 2016-01-20 | | Last modified: | 2016-01-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Ebola Viral Glycoprotein Bound to Its Endosomal Receptor Niemann-Pick C1.
Cell, 164, 2016
|
|
5W2H
 
 | | Crystal structure of the core catalytic domain of human inositol phosphate multikinase in complex with Ins(1,4,5)P3 and ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, D-MYO-INOSITOL-1,4,5-TRIPHOSPHATE, Inositol polyphosphate multikinase,Inositol polyphosphate multikinase, ... | | Authors: | Wang, H, Shears, S.B. | | Deposit date: | 2017-06-06 | | Release date: | 2017-09-13 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural features of human inositol phosphate multikinase rationalize its inositol phosphate kinase and phosphoinositide 3-kinase activities.
J. Biol. Chem., 292, 2017
|
|
5F1B
 
 | | Structural basis of Ebola virus entry: viral glycoprotein bound to its endosomal receptor Niemann-Pick C1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, GP1, GP2, ... | | Authors: | Wang, H, Shi, Y, Song, J, Qi, J, Lu, G, Yan, J, Gao, G.F. | | Deposit date: | 2015-11-30 | | Release date: | 2016-01-20 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Ebola Viral Glycoprotein Bound to Its Endosomal Receptor Niemann-Pick C1.
Cell, 164, 2016
|
|
5B6O
 
 | | Crystal structure of MS8104 | | Descriptor: | 3C-like proteinase | | Authors: | Wang, H, Kim, Y, Muramatsu, T, Takemoto, C, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2016-05-31 | | Release date: | 2016-06-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.202 Å) | | Cite: | SARS-CoV 3CL protease cleaves its C-terminal autoprocessing site by novel subsite cooperativity
Proc. Natl. Acad. Sci. U.S.A., 113, 2016
|
|
7YRN
 
 | |
1BPR
 
 | | NMR STRUCTURE OF THE SUBSTRATE BINDING DOMAIN OF DNAK, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | DNAK | | Authors: | Wang, H, Kurochkin, A.V, Pang, Y, Hu, W, Flynn, G.C, Zuiderweg, E.R.P. | | Deposit date: | 1998-08-11 | | Release date: | 1999-03-02 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | NMR solution structure of the 21 kDa chaperone protein DnaK substrate binding domain: a preview of chaperone-protein interaction.
Biochemistry, 37, 1998
|
|
4ED5
 
 | | Crystal structure of the two N-terminal RRM domains of HuR complexed with RNA | | Descriptor: | 1,2-ETHANEDIOL, 1-METHOXY-2-(2-METHOXYETHOXY)ETHANE, 5'-R(*A*UP*UP*UP*UP*UP*AP*UP*UP*UP*U)-3', ... | | Authors: | Wang, H, Zeng, F, Liu, Q, Niu, L, Teng, M, Li, X. | | Deposit date: | 2012-03-27 | | Release date: | 2012-05-23 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The structure of the ARE-binding domains of Hu antigen R (HuR) undergoes conformational changes during RNA binding.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
3V94
 
 | | TcrPDEC1 catalytic domain in complex with inhibitor wyq16 | | Descriptor: | Cyclic nucleotide specific phosphodiesterase, MAGNESIUM ION, ZINC ION, ... | | Authors: | Wang, H, Kunz, S, Chen, G, Seebeck, T, Wan, Y, Robinson, H, Martinelli, S, Ke, H. | | Deposit date: | 2011-12-23 | | Release date: | 2012-02-22 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | TcrPDEC1 catalytic domain in complex with inhibitor wyq16
To be Published
|
|
2NZ0
 
 | | Crystal structure of potassium channel Kv4.3 in complex with its regulatory subunit KChIP1 | | Descriptor: | CALCIUM ION, Kv channel-interacting protein 1, Potassium voltage-gated channel subfamily D member 3, ... | | Authors: | Wang, H, Yan, Y, Shen, Y, Chen, L, Wang, K. | | Deposit date: | 2006-11-22 | | Release date: | 2006-12-26 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural basis for modulation of Kv4 K(+) channels by auxiliary KChIP subunits.
Nat.Neurosci., 10, 2007
|
|
5ZM8
 
 | | Crystal structure of ORP2-ORD in complex with PI(4,5)P2 | | Descriptor: | Oxysterol-binding protein-related protein 2, [(2~{S})-1-octadecanoyloxy-3-[oxidanyl-[(1~{R},2~{R},3~{S},4~{S},5~{S},6~{S})-2,3,6-tris(oxidanyl)-4,5-diphosphonooxy-cyclohexyl]oxy-phosphoryl]oxy-propan-2-yl] icosa-5,8,11,14-tetraenoate | | Authors: | Wang, H, Dong, J.Q, Wang, J, Wu, J.W. | | Deposit date: | 2018-04-01 | | Release date: | 2019-01-02 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | ORP2 Delivers Cholesterol to the Plasma Membrane in Exchange for Phosphatidylinositol 4, 5-Bisphosphate (PI(4,5)P2).
Mol. Cell, 73, 2019
|
|
7ZSB
 
 | |
7ZSA
 
 | |
7ZS9
 
 | |
