2RIE
 
 | | Crystal structure of the trimeric neck and carbohydrate recognition domain of human surfactant protein D in complex with 2-deoxy-L-glycero-D-manno-heptose | | Descriptor: | 2-deoxy-beta-L-galacto-heptopyranose, CALCIUM ION, Pulmonary surfactant-associated protein D | | Authors: | Wang, H, Head, J, Kosma, P, Sheikh, S, McDonald, B, Smith, K, Cafarella, T, Seaton, B, Crouch, E. | | Deposit date: | 2007-10-10 | | Release date: | 2008-01-15 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Recognition of heptoses and the inner core of bacterial lipopolysaccharides by surfactant protein d.
Biochemistry, 47, 2008
|
|
2DUC
 
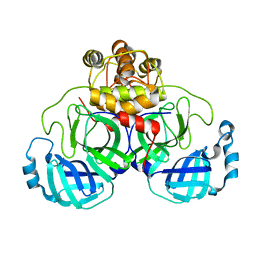 | | Crystal structure of SARS coronavirus main proteinase(3CLPRO) | | Descriptor: | Replicase polyprotein 1ab | | Authors: | Wang, H, Kim, Y.T, Muramatsu, T, Takemoto, C, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2006-07-21 | | Release date: | 2007-07-24 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | SARS-CoV 3CL protease cleaves its C-terminal autoprocessing site by novel subsite cooperativity
Proc. Natl. Acad. Sci. U.S.A., 113, 2016
|
|
8HZZ
 
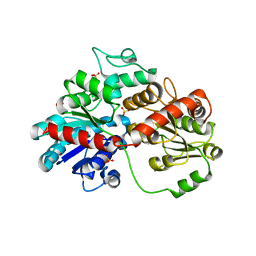 | | GuApiGT (UGT79B74) | | Descriptor: | SULFATE ION, apiosyltransferase | | Authors: | Wang, H.T, Wang, Z.L, Li, F.D, Ye, M. | | Deposit date: | 2023-01-10 | | Release date: | 2023-09-20 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Insights into the missing apiosylation step in flavonoid apiosides biosynthesis of Leguminosae plants.
Nat Commun, 14, 2023
|
|
8I0E
 
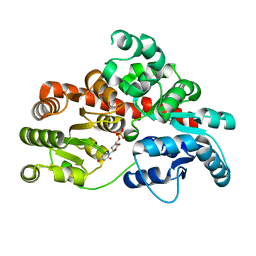 | | Sb3GT1 complex with UDP | | Descriptor: | Glycosyltransferase, URIDINE-5'-DIPHOSPHATE | | Authors: | Wang, H.T, Wang, Z.L, Ye, M. | | Deposit date: | 2023-01-10 | | Release date: | 2023-09-20 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Insights into the missing apiosylation step in flavonoid apiosides biosynthesis of Leguminosae plants.
Nat Commun, 14, 2023
|
|
8I0D
 
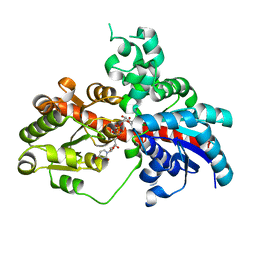 | | Sb3GT1 375S/Q377H mutant complex with UDP-Glc | | Descriptor: | Glycosyltransferase, URIDINE-5'-DIPHOSPHATE-GLUCOSE | | Authors: | Wang, H.T, Wang, Z.L, Ye, M. | | Deposit date: | 2023-01-10 | | Release date: | 2023-09-20 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | Insights into the missing apiosylation step in flavonoid apiosides biosynthesis of Leguminosae plants.
Nat Commun, 14, 2023
|
|
3K3E
 
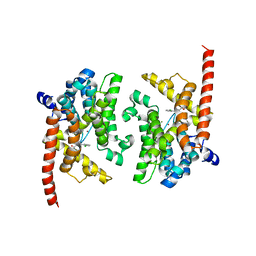 | | Crystal structure of the PDE9A catalytic domain in complex with (R)-BAY73-6691 | | Descriptor: | 1-(2-chlorophenyl)-6-[(2R)-3,3,3-trifluoro-2-methylpropyl]-1,7-dihydro-4H-pyrazolo[3,4-d]pyrimidin-4-one, High affinity cGMP-specific 3',5'-cyclic phosphodiesterase 9A, MAGNESIUM ION, ... | | Authors: | Wang, H, Luo, X, Ye, M, Hou, J, Robinson, H, Ke, H. | | Deposit date: | 2009-10-02 | | Release date: | 2010-02-16 | | Last modified: | 2013-11-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Insight into Binding of Phosphodiesterase-9A Selective Inhibitors by Crystal Structures and Mutagenesis
J.Med.Chem., 53, 2010
|
|
3K3H
 
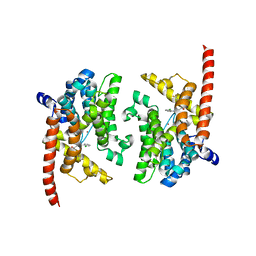 | | Crystal structure of the PDE9A catalytic domain in complex with (S)-BAY73-6691 | | Descriptor: | 1-(2-chlorophenyl)-6-[(2S)-3,3,3-trifluoro-2-methylpropyl]-1,7-dihydro-4H-pyrazolo[3,4-d]pyrimidin-4-one, High affinity cGMP-specific 3',5'-cyclic phosphodiesterase 9A, MAGNESIUM ION, ... | | Authors: | Wang, H, Luo, X, Ye, M, Hou, J, Robinson, H, Ke, H. | | Deposit date: | 2009-10-02 | | Release date: | 2010-02-16 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Insight into Binding of Phosphodiesterase-9A Selective Inhibitors by Crystal Structures and Mutagenesis
J.Med.Chem., 53, 2010
|
|
3WDG
 
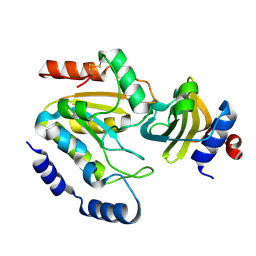 | | Staphylococcus aureus UDG / UGI complex | | Descriptor: | Uncharacterized protein, Uracil-DNA glycosylase | | Authors: | Wang, H.C, Ko, T.P, Wang, A.H.J. | | Deposit date: | 2013-06-18 | | Release date: | 2014-02-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Staphylococcus aureus protein SAUGI acts as a uracil-DNA glycosylase inhibitor.
Nucleic Acids Res., 42, 2013
|
|
3WDF
 
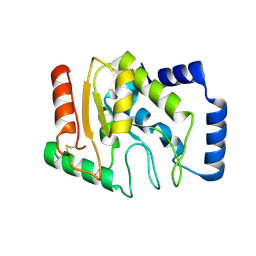 | | Staphylococcus aureus UDG | | Descriptor: | Uracil-DNA glycosylase | | Authors: | Wang, H.C, Ko, T.P, Wang, A.H.J. | | Deposit date: | 2013-06-18 | | Release date: | 2014-02-05 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Staphylococcus aureus protein SAUGI acts as a uracil-DNA glycosylase inhibitor.
Nucleic Acids Res., 42, 2013
|
|
6VCD
 
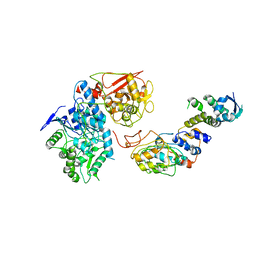 | | Cryo-EM structure of IRP2-FBXL5-SKP1 complex | | Descriptor: | F-box/LRR-repeat protein 5, FE2/S2 (INORGANIC) CLUSTER, Iron-responsive element binding protein 2, ... | | Authors: | Wang, H, Shi, H, Zheng, N. | | Deposit date: | 2019-12-20 | | Release date: | 2020-08-05 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | FBXL5 Regulates IRP2 Stability in Iron Homeostasis via an Oxygen-Responsive [2Fe2S] Cluster.
Mol.Cell, 78, 2020
|
|
1BM5
 
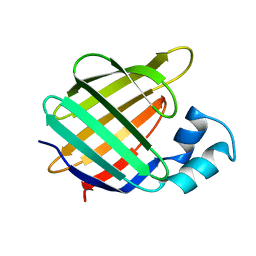 | |
6LHE
 
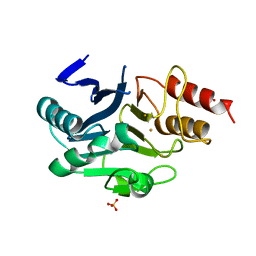 | | Crystal Structure of Gold-bound NDM-1 | | Descriptor: | GOLD ION, Metallo-beta-lactamase type 2, SULFATE ION | | Authors: | Wang, H, Sun, H, Wang, M. | | Deposit date: | 2019-12-07 | | Release date: | 2020-09-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.206 Å) | | Cite: | Resensitizing carbapenem- and colistin-resistant bacteria to antibiotics using auranofin.
Nat Commun, 11, 2020
|
|
8R0F
 
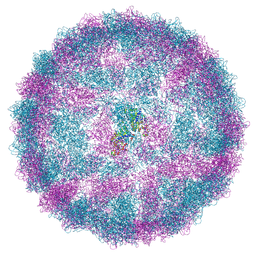 | | Capsid structure of Giardiavirus (GLV) HP strain | | Descriptor: | Capsid protein | | Authors: | Wang, H, Gianluca, M, Munke, A, Hassan, M.M, Lalle, M, Okamoto, K. | | Deposit date: | 2023-10-31 | | Release date: | 2024-04-03 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.14 Å) | | Cite: | High-resolution comparative atomic structures of two Giardiavirus prototypes infecting G. duodenalis parasite.
Plos Pathog., 20, 2024
|
|
8R0G
 
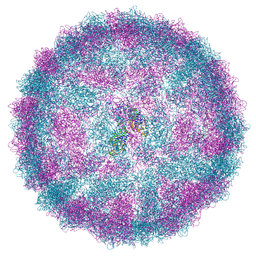 | | Capsid structure of Giardiavirus (GLV) CAT strain | | Descriptor: | Capsid protein | | Authors: | Wang, H, Gianluca, M, Munke, A, Hassan, M.M, Lalle, M, Okamoto, K. | | Deposit date: | 2023-10-31 | | Release date: | 2024-04-03 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | High-resolution comparative atomic structures of two Giardiavirus prototypes infecting G. duodenalis parasite.
Plos Pathog., 20, 2024
|
|
3KUZ
 
 | | Crystal structure of the ubiquitin like domain of PLXNC1 | | Descriptor: | Plexin-C1, UNKNOWN ATOM OR ION | | Authors: | Wang, H, Li, B, Tempel, W, Tong, Y, Guan, X, Zhong, N, Crombet, L, MacKenzie, F, Buck, M, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Weigelt, J, Bochkarev, A, Park, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-11-28 | | Release date: | 2009-12-08 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of the ubiquitin like domain of PLXNC1
to be published
|
|
3ECN
 
 | | Crystal structure of PDE8A catalytic domain in complex with IBMX | | Descriptor: | 3-ISOBUTYL-1-METHYLXANTHINE, High affinity cAMP-specific and IBMX-insensitive 3',5'-cyclic phosphodiesterase 8A, MAGNESIUM ION, ... | | Authors: | Wang, H, Yan, Z, Yang, S, Cai, J, Robinson, H, Ke, H. | | Deposit date: | 2008-09-01 | | Release date: | 2008-11-25 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Kinetic and structural studies of phosphodiesterase-8A and implication on the inhibitor selectivity
Biochemistry, 47, 2008
|
|
3ECM
 
 | | Crystal structure of the unliganded PDE8A catalytic domain | | Descriptor: | High affinity cAMP-specific and IBMX-insensitive 3',5'-cyclic phosphodiesterase 8A, MAGNESIUM ION, ZINC ION | | Authors: | Wang, H, Yan, Z, Yang, S, Cai, J, Robinson, H, Ke, H. | | Deposit date: | 2008-09-01 | | Release date: | 2008-11-25 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Kinetic and structural studies of phosphodiesterase-8A and implication on the inhibitor selectivity
Biochemistry, 47, 2008
|
|
6N5C
 
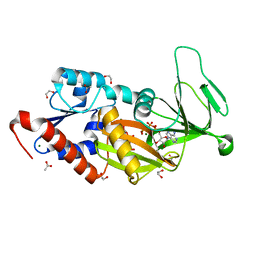 | | Crystal structure of the catalytic domain of PPIP5K2 in complex with AMPPNP and 5-PCF2Am-InsP5 | | Descriptor: | (1,1-difluoro-2-oxo-2-{[(1s,2R,3S,4s,5R,6S)-2,3,4,5,6-pentakis(phosphonooxy)cyclohexyl]amino}ethyl)phosphonic acid, 1,2-ETHANEDIOL, ACETATE ION, ... | | Authors: | Wang, H, Shears, S.B, Riley, A, Potter, B. | | Deposit date: | 2018-11-21 | | Release date: | 2019-08-21 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Synthesis of an alpha-phosphono-alpha , alpha-difluoroacetamide analogue of the diphosphoinositol pentakisphosphate 5-InsP7.
Medchemcomm, 10, 2019
|
|
3LFV
 
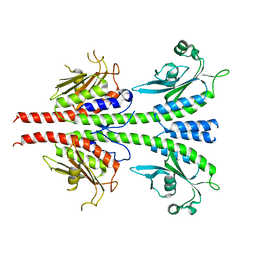 | | crystal structure of unliganded PDE5A GAF domain | | Descriptor: | cGMP-specific 3',5'-cyclic phosphodiesterase | | Authors: | Wang, H, Robinson, H, Ke, H. | | Deposit date: | 2010-01-18 | | Release date: | 2010-09-22 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Conformation changes, N-terminal involvement, and cGMP signal relay in the phosphodiesterase-5 GAF domain.
J.Biol.Chem., 285, 2010
|
|
5YQM
 
 | | Crystal structure of Sirt2 in complex with selective inhibitor A29 | | Descriptor: | 2-(4,6-dimethylpyrimidin-2-yl)sulfanyl-N-(4-phenylsulfanylphenyl)ethanamide, BETA-MERCAPTOETHANOL, NAD-dependent protein deacetylase sirtuin-2, ... | | Authors: | Wang, H, Yu, Y, Li, G, chen, Q. | | Deposit date: | 2017-11-07 | | Release date: | 2018-10-17 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.735 Å) | | Cite: | X-ray crystal structure guided discovery of new selective, substrate-mimicking sirtuin 2 inhibitors that exhibit activities against non-small cell lung cancer cells.
Eur J Med Chem, 155, 2018
|
|
3MF0
 
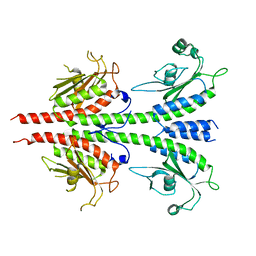 | | Crystal structure of PDE5A GAF domain (89-518) | | Descriptor: | cGMP-specific 3',5'-cyclic phosphodiesterase | | Authors: | Wang, H, Robinson, H, Ke, H. | | Deposit date: | 2010-04-01 | | Release date: | 2010-09-22 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Conformation changes, N-terminal involvement, and cGMP signal relay in the phosphodiesterase-5 GAF domain.
J.Biol.Chem., 285, 2010
|
|
7U2L
 
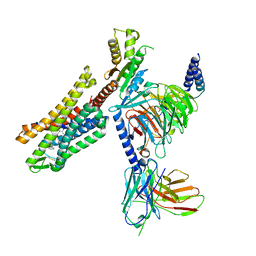 | | C5guano-uOR-Gi-scFv16 | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(i) subunit alpha-1, ... | | Authors: | Wang, H, Qu, Q, Skiniotis, G, Kobilka, B. | | Deposit date: | 2022-02-24 | | Release date: | 2022-05-04 | | Last modified: | 2023-02-08 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structure-based design of bitopic ligands for the μ-opioid receptor.
Nature, 613, 2023
|
|
3O47
 
 | | Crystal structure of ARFGAP1-ARF1 fusion protein | | Descriptor: | ADP-ribosylation factor GTPase-activating protein 1, ADP-ribosylation factor 1, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Wang, H, Tong, Y, Nedyalkova, L, Tempel, W, Guan, X, Crombet, L, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Weigelt, J, Bochkarev, A, Park, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-07-26 | | Release date: | 2010-08-11 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of ARFGAP1-ARF1 fusion protein
to be published
|
|
5YQN
 
 | | Crystal structure of Sirt2 in complex with selective inhibitor L55 | | Descriptor: | DI(HYDROXYETHYL)ETHER, N-[3-[[3-[2-(4,6-dimethylpyrimidin-2-yl)sulfanylethanoylamino]phenyl]methoxy]phenyl]-1-methyl-pyrazole-4-carboxamide, NAD-dependent protein deacetylase sirtuin-2, ... | | Authors: | Wang, H, Yu, Y, Li, G, Chen, Q. | | Deposit date: | 2017-11-07 | | Release date: | 2018-10-17 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | X-ray crystal structure guided discovery of new selective, substrate-mimicking sirtuin 2 inhibitors that exhibit activities against non-small cell lung cancer cells.
Eur J Med Chem, 155, 2018
|
|
5YQO
 
 | | Crystal structure of Sirt2 in complex with selective inhibitor L5C | | Descriptor: | N-[4-[[3-[2-(4,6-dimethylpyrimidin-2-yl)sulfanylethanoylamino]phenyl]methoxy]phenyl]-1-methyl-pyrazole-4-carboxamide, NAD-dependent protein deacetylase sirtuin-2, ZINC ION | | Authors: | Wang, H, Yu, Y, Li, G, Chen, Q. | | Deposit date: | 2017-11-07 | | Release date: | 2018-10-17 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.483 Å) | | Cite: | X-ray crystal structure guided discovery of new selective, substrate-mimicking sirtuin 2 inhibitors that exhibit activities against non-small cell lung cancer cells.
Eur J Med Chem, 155, 2018
|
|
