3GTJ
 
 | | Backtracked RNA polymerase II complex with 13mer RNA | | Descriptor: | DNA (28-MER), DNA (5'-D(*CP*TP*GP*CP*TP*TP*AP*TP*CP*GP*GP*TP*AP*G)-3'), DNA-directed RNA polymerase II subunit RPB1, ... | | Authors: | Wang, D, Bushnell, D.A, Huang, X, Westover, K.D, Levitt, M, Kornberg, R.D. | | Deposit date: | 2009-03-27 | | Release date: | 2009-06-09 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.42 Å) | | Cite: | Structural basis of transcription: backtracked RNA polymerase II at 3.4 angstrom resolution.
Science, 324, 2009
|
|
5EYO
 
 | | The crystal structure of the Max bHLH domain in complex with 5-carboxyl cytosine DNA | | Descriptor: | DNA (5'-D(*AP*GP*TP*AP*GP*CP*AP*(1CC)P*GP*TP*GP*CP*TP*AP*CP*T)-3'), Protein max | | Authors: | Wang, D, Hashimoto, H, Zhang, X, Cheng, X. | | Deposit date: | 2015-11-25 | | Release date: | 2016-12-14 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | MAX is an epigenetic sensor of 5-carboxylcytosine and is altered in multiple myeloma.
Nucleic Acids Res., 45, 2017
|
|
8JED
 
 | | Crystal structure of mRNA cap (guanine-N7) methyltransferase E12 subunit from monkeypox virus and discovery of its inhibitors | | Descriptor: | mRNA-capping enzyme regulatory subunit OPG124 | | Authors: | Wang, D, Zhao, R, Shu, W, Hu, W, Wang, M, Cao, J, Zhou, X. | | Deposit date: | 2023-05-15 | | Release date: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Crystal structure of mRNA cap (guanine-N7) methyltransferase E12 subunit from monkeypox virus and discovery of its inhibitors.
Int.J.Biol.Macromol., 253, 2023
|
|
5GSZ
 
 | | Crystal Structure of the KIF19A Motor Domain Complexed with Mg-ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Kinesin-like protein KIF19, MAGNESIUM ION | | Authors: | Wang, D, Nitta, R, Hirokawa, N. | | Deposit date: | 2016-08-18 | | Release date: | 2016-09-28 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.72 Å) | | Cite: | Motility and microtubule depolymerization mechanisms of the Kinesin-8 motor, KIF19A
Elife, 5, 2016
|
|
5H2T
 
 | |
5GPO
 
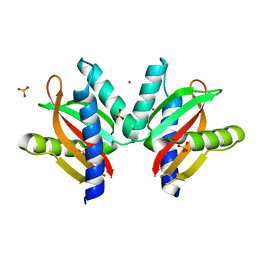 | | The sensor domain structure of the zinc-responsive histidine kinase CzcS from Pseudomonas Aeruginosa | | Descriptor: | SULFATE ION, Sensor protein CzcS, ZINC ION | | Authors: | Wang, D, Chen, W.Z, Huang, S.Q, Liu, X.C, Hu, Q.Y, Wei, T.B, Gan, J.H, Chen, H. | | Deposit date: | 2016-08-03 | | Release date: | 2017-08-09 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.701 Å) | | Cite: | Structural basis of Zn(II) induced metal detoxification and antibiotic resistance by histidine kinase CzcS in Pseudomonas aeruginosa
PLoS Pathog., 13, 2017
|
|
4FFJ
 
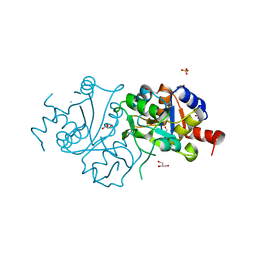 | | The crystal structure of spDHBPs from S.pneumoniae | | Descriptor: | GLYCEROL, Riboflavin biosynthesis protein ribBA, SULFATE ION | | Authors: | Wang, D. | | Deposit date: | 2012-06-01 | | Release date: | 2013-07-17 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | The crystal structure of SpDHBPs from S. pneumoniae
To be Published
|
|
2NVQ
 
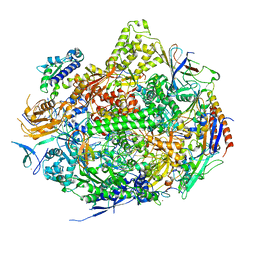 | | RNA Polymerase II Elongation Complex in 150 mM Mg+2 with 2'dUTP | | Descriptor: | 28-MER DNA template strand, 5'-D(*CP*TP*GP*CP*TP*TP*AP*TP*CP*GP*GP*TP*AP*G)-3', 5'-R(*AP*UP*CP*GP*AP*GP*AP*GP*GP*A)-3', ... | | Authors: | Wang, D, Bushnell, D.A, Westover, K.D, Kaplan, C.D, Kornberg, R.D. | | Deposit date: | 2006-11-13 | | Release date: | 2006-12-12 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural basis of transcription: role of the trigger loop in substrate specificity and catalysis
Cell(Cambridge,Mass.), 127, 2006
|
|
2NVT
 
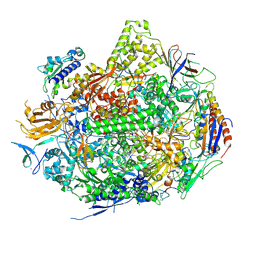 | | RNA Polymerase II Elongation Complex in 150 mM Mg+2 with GMPCPP | | Descriptor: | 5'-D(*GP*TP*AP*CP*TP*TP*G)-3', 5'-D(P*CP*AP*AP*GP*TP*AP*CP*TP*TP*AP*CP*GP*CP*CP*TP*GP*GP*TP*CP*TP*T)-3', 5'-R(*AP*AP*GP*AP*CP*CP*AP*GP*GP*C)-3', ... | | Authors: | Wang, D, Bushnell, D.A, Westover, K.D, Kaplan, C.D, Kornberg, R.D. | | Deposit date: | 2006-11-13 | | Release date: | 2006-12-12 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.36 Å) | | Cite: | Structural basis of transcription: role of the trigger loop in substrate specificity and catalysis
Cell(Cambridge,Mass.), 127, 2006
|
|
2NVZ
 
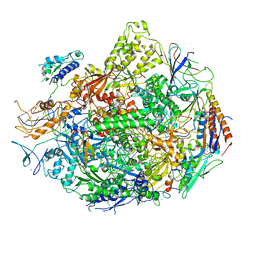 | | RNA Polymerase II elongation complex with UTP, updated 11/2006 | | Descriptor: | 28-MER DNA template strand, 5'-D(*CP*TP*GP*CP*TP*TP*AP*TP*CP*GP*GP*TP*AP*G)-3', 5'-R(*AP*UP*CP*GP*AP*GP*AP*GP*GP*A)-3', ... | | Authors: | Wang, D, Bushnell, D.A, Westover, K.D, Kaplan, C.D, Kornberg, R.D. | | Deposit date: | 2006-11-14 | | Release date: | 2006-12-19 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (4.3 Å) | | Cite: | Structural basis of transcription: role of the trigger loop in substrate specificity and catalysis
Cell(Cambridge,Mass.), 127, 2006
|
|
2NVX
 
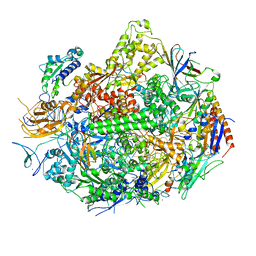 | | RNA polymerase II elongation complex in 5 mM Mg+2 with 2'-dUTP | | Descriptor: | 28-MER DNA template strand, 5'-D(*CP*TP*GP*CP*TP*TP*AP*TP*CP*GP*GP*TP*AP*G)-3', 5'-R(*AP*UP*CP*GP*AP*GP*AP*GP*GP*A)-3', ... | | Authors: | Wang, D, Bushnell, D.A, Westover, K.D, Kaplan, C.D, Kornberg, R.D. | | Deposit date: | 2006-11-13 | | Release date: | 2006-12-19 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Structural basis of transcription: role of the trigger loop in substrate specificity and catalysis
Cell(Cambridge,Mass.), 127, 2006
|
|
2NVY
 
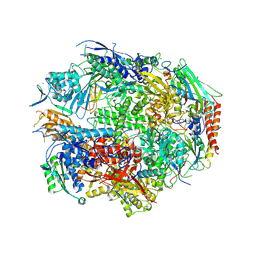 | | RNA Polymerase II form II in 150 mM Mn+2 | | Descriptor: | DNA-directed RNA polymerase II 13.6 kDa polypeptide, DNA-directed RNA polymerase II 140 kDa polypeptide, DNA-directed RNA polymerase II 45 kDa polypeptide, ... | | Authors: | Wang, D, Bushnell, D.A, Westover, K.D, Kaplan, C.D, Kornberg, R.D. | | Deposit date: | 2006-11-13 | | Release date: | 2006-12-26 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Structural basis of transcription: role of the trigger loop in substrate specificity and catalysis
Cell(Cambridge,Mass.), 127, 2006
|
|
7F4A
 
 | |
7F3S
 
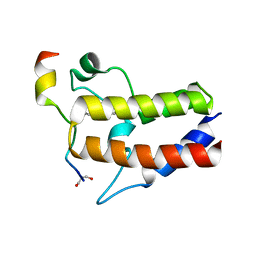 | |
7F51
 
 | | Crystal structure of Hst2 in complex with 2'-O-Benzoyl ADP Ribose | | Descriptor: | NAD-dependent protein deacetylase HST2, ZINC ION, [(2R,3R,4R,5R)-5-[[[[(2R,3S,4R,5R)-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methoxy-oxidanyl-phosphoryl]oxy-oxidanyl-phosphoryl]oxymethyl]-2,4-bis(oxidanyl)oxolan-3-yl] benzoate | | Authors: | Wang, D, Yan, F, Chen, Y. | | Deposit date: | 2021-06-21 | | Release date: | 2022-03-16 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Global profiling of regulatory elements in the histone benzoylation pathway.
Nat Commun, 13, 2022
|
|
7F4E
 
 | |
7F5M
 
 | |
1W24
 
 | | Crystal Structure Of human Vps29 | | Descriptor: | VACUOLAR PROTEIN SORTING PROTEIN 29 | | Authors: | Wang, D, Guo, M, Teng, M, Niu, L. | | Deposit date: | 2004-06-26 | | Release date: | 2005-03-23 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structure of Human Vacuolar Protein Sorting Protein 29 Reveals a Phosphodiesterase/Nuclease-Like Fold and Two Protein-Protein Interaction Sites.
J.Biol.Chem., 280, 2005
|
|
7V7C
 
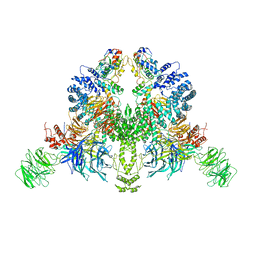 | | CryoEM structure of DDB1-VprBP-Vpr-UNG2(94-313) complex | | Descriptor: | DDB1- and CUL4-associated factor 1, DNA damage-binding protein 1, Protein Vpr, ... | | Authors: | Wang, D, Xu, J, Liu, Q, Xiang, Y. | | Deposit date: | 2021-08-21 | | Release date: | 2022-08-31 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structural insights into the HIV-1 Vpr mediated ubiquitination through the Cullin-RING E3 ubiquitin ligase
To Be Published
|
|
7V7B
 
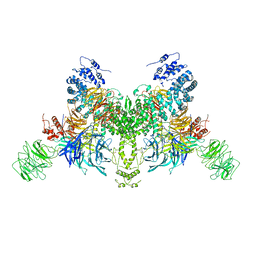 | |
4X67
 
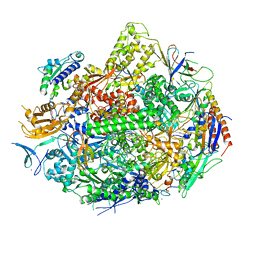 | | Crystal structure of elongating yeast RNA polymerase II stalled at oxidative Cyclopurine DNA lesions. | | Descriptor: | DNA-directed RNA polymerase II subunit RPB1, DNA-directed RNA polymerase II subunit RPB11, DNA-directed RNA polymerase II subunit RPB2, ... | | Authors: | Wang, L, Chong, J, Wang, D. | | Deposit date: | 2014-12-07 | | Release date: | 2015-02-04 | | Last modified: | 2019-12-25 | | Method: | X-RAY DIFFRACTION (4.1 Å) | | Cite: | Mechanism of RNA polymerase II bypass of oxidative cyclopurine DNA lesions.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
4KOM
 
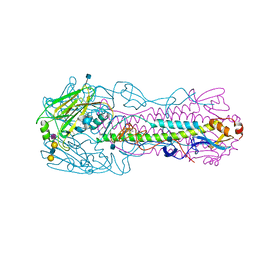 | | The structure of hemagglutinin from avian-origin H7N9 influenza virus in complex with avian receptor analog 3'SLNLN (NeuAcα2-3Galβ1-4GlcNAcβ1-3Galβ1-4Glc) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin HA1, Hemagglutinin HA2, ... | | Authors: | Shi, Y, Zhang, W, Wang, F, Qi, J, Song, H, Wu, Y, Gao, F, Zhang, Y, Fan, Z, Gong, W, Wang, D, Shu, Y, Wang, Y, Yan, J, Gao, G.F. | | Deposit date: | 2013-05-12 | | Release date: | 2013-11-06 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.604 Å) | | Cite: | Structures and receptor binding of hemagglutinins from human-infecting H7N9 influenza viruses.
Science, 342, 2013
|
|
4KON
 
 | | The structure of hemagglutinin from avian-origin H7N9 influenza virus in complex with human receptor analog 6'SLNLN (NeuAcα2-6Galβ1-4GlcNAcβ1-3Galβ1-4Glc) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin HA1, Hemagglutinin HA2, ... | | Authors: | Shi, Y, Zhang, W, Wang, F, Qi, J, Song, H, Wu, Y, Gao, F, Zhang, Y, Fan, Z, Gong, W, Wang, D, Shu, Y, Wang, Y, Yan, J, Gao, G.F. | | Deposit date: | 2013-05-12 | | Release date: | 2013-11-06 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structures and receptor binding of hemagglutinins from human-infecting H7N9 influenza viruses.
Science, 342, 2013
|
|
4KOL
 
 | | The structure of hemagglutinin from avian-origin H7N9 influenza virus | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin HA1, Hemagglutinin HA2 | | Authors: | Shi, Y, Zhang, W, Wang, F, Qi, J, Song, H, Wu, Y, Gao, F, Zhang, Y, Fan, Z, Gong, W, Wang, D, Shu, Y, Wang, Y, Yan, J, Gao, G.F. | | Deposit date: | 2013-05-12 | | Release date: | 2013-11-06 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.799 Å) | | Cite: | Structures and receptor binding of hemagglutinins from human-infecting H7N9 influenza viruses.
Science, 342, 2013
|
|
4UML
 
 | | Crystal structure of ganglioside induced differentiation associated protein 2 (GDAP2) macro domain | | Descriptor: | GANGLIOSIDE-INDUCED DIFFERENTIATION-ASSOCIATED PROTEIN 2 | | Authors: | Elkins, J.M, Wang, J, Kopec, J, Wang, D, Strain-Damerell, C, Shrestha, L, Sieg, C, Tallant, C, Newman, J.A, von Delft, F, Bountra, C, Edwards, A, Knapp, S. | | Deposit date: | 2014-05-19 | | Release date: | 2014-05-28 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of Gdap2
To be Published
|
|
