9O4A
 
 | | Ec83 Retron PtuAB mutant complex | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Retron Ec83 probable ATPase, Retron Ec83 putative HNH endonuclease, ... | | Authors: | Wang, C, Rish, A, Fu, T.M. | | Deposit date: | 2025-04-08 | | Release date: | 2025-07-02 | | Last modified: | 2025-09-03 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Disassembly activates Retron-Septu for antiphage defense.
Science, 389, 2025
|
|
7WN0
 
 | | Structure of PfENT1(Y190A) in complex with nanobody 19 | | Descriptor: | Equilibrative nucleoside/nucleobase transporter, nanobody19 | | Authors: | Wang, C, Deng, D, Ren, R.B, Yu, L.Y. | | Deposit date: | 2022-01-17 | | Release date: | 2023-02-01 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.64 Å) | | Cite: | Structural basis of the substrate recognition and inhibition mechanism of Plasmodium falciparum nucleoside transporter PfENT1.
Nat Commun, 14, 2023
|
|
7WN1
 
 | | Structure of PfNT1(Y190A) in complex with nanobody 48 and inosine | | Descriptor: | Equilibrative nucleoside/nucleobase transporter, INOSINE, nanobody48 | | Authors: | Wang, C, Deng, D, Ren, R.B, Yu, L.Y. | | Deposit date: | 2022-01-17 | | Release date: | 2023-02-01 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.11 Å) | | Cite: | Structural basis of the substrate recognition and inhibition mechanism of Plasmodium falciparum nucleoside transporter PfENT1.
Nat Commun, 14, 2023
|
|
1F0Z
 
 | | SOLUTION STRUCTURE OF THIS, THE SULFUR CARRIER PROTEIN IN E.COLI THIAMIN BIOSYNTHESIS | | Descriptor: | THIS PROTEIN | | Authors: | Wang, C, Xi, J, Begley, T.P, Nicholson, L.K. | | Deposit date: | 2000-05-17 | | Release date: | 2001-01-10 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of ThiS and implications for the evolutionary roots of ubiquitin.
Nat.Struct.Biol., 8, 2001
|
|
5XZ7
 
 | | Crystal Structure of Phosphofructokinase from Staphylococcus aureus in complex with adenylylimidodiphosphate, the ATP analogue | | Descriptor: | 6-O-phosphono-beta-D-fructofuranose, ATP-dependent 6-phosphofructokinase, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Wang, C.L, Tian, T, Zang, J.Y. | | Deposit date: | 2017-07-11 | | Release date: | 2019-03-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural Insights into the Regulation of Staphylococcus aureus Phosphofructokinase by Tetramer-Dimer Conversion.
Biochemistry, 57, 2018
|
|
5XZ8
 
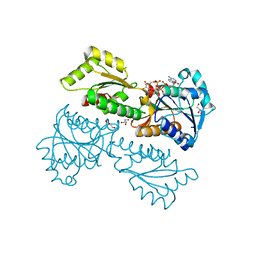 | | Crystal Structure of Phosphofructokinase from Staphylococcus aureus in complex with adenylylimidodiphosphate (the ATP analog) and fructose-6-phosphate | | Descriptor: | 6-O-phosphono-beta-D-fructofuranose, ATP-dependent 6-phosphofructokinase, GLYCEROL, ... | | Authors: | Wang, C.L, Tian, T, Zang, J.Y. | | Deposit date: | 2017-07-11 | | Release date: | 2019-02-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural Insights into the Regulation of Staphylococcus aureus Phosphofructokinase by Tetramer-Dimer Conversion.
Biochemistry, 57, 2018
|
|
5XZ6
 
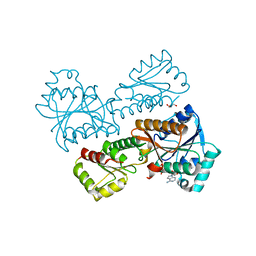 | | Crystal Structure of Phosphofructokinase from Staphylococcus aureus in complex with adenylylimidodiphosphate, the ATP analogue | | Descriptor: | ATP-dependent 6-phosphofructokinase, GLYCEROL, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER | | Authors: | Wang, C.L, Tian, T, Zang, J.Y. | | Deposit date: | 2017-07-11 | | Release date: | 2019-02-27 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural Insights into the Regulation of Staphylococcus aureus Phosphofructokinase by Tetramer-Dimer Conversion.
Biochemistry, 57, 2018
|
|
5XZA
 
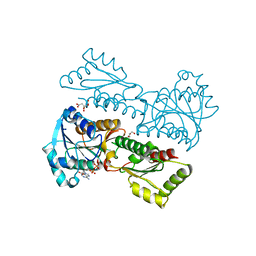 | | Crystal Structure of Phosphofructokinase from Staphylococcus aureus in complex with ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP-dependent 6-phosphofructokinase, CITRATE ANION, ... | | Authors: | Wang, C.L, Tian, T, Zang, J.Y. | | Deposit date: | 2017-07-12 | | Release date: | 2019-03-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Insights into the Regulation of Staphylococcus aureus Phosphofructokinase by Tetramer-Dimer Conversion.
Biochemistry, 57, 2018
|
|
5Y5W
 
 | |
5E8L
 
 | | Crystal structure of geranylgeranyl pyrophosphate synthase 11 from Arabidopsis thaliana | | Descriptor: | Heterodimeric geranylgeranyl pyrophosphate synthase large subunit 1, chloroplastic | | Authors: | Wang, C, Chen, Q, Fan, D, Li, J, Wang, G, Zhang, P. | | Deposit date: | 2015-10-14 | | Release date: | 2015-11-11 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.807 Å) | | Cite: | Structural Analyses of Short-Chain Prenyltransferases Identify an Evolutionarily Conserved GFPPS Clade in Brassicaceae Plants.
Mol Plant, 9, 2016
|
|
5E8K
 
 | | Crystal structure of polyprenyl pyrophosphate synthase 2 from Arabidopsis thaliana | | Descriptor: | Geranylgeranyl pyrophosphate synthase 10, mitochondrial | | Authors: | Wang, C, Chen, Q, Fan, D, Li, J, Wang, G, Zhang, P. | | Deposit date: | 2015-10-14 | | Release date: | 2015-11-11 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3.028 Å) | | Cite: | Structural Analyses of Short-Chain Prenyltransferases Identify an Evolutionarily Conserved GFPPS Clade in Brassicaceae Plants.
Mol Plant, 9, 2016
|
|
5E8H
 
 | | Crystal structure of geranylfarnesyl pyrophosphate synthases 2 from Arabidopsis thaliana | | Descriptor: | Geranylgeranyl pyrophosphate synthase 3, chloroplastic | | Authors: | Wang, C, Chen, Q, Wang, G, Zhang, P. | | Deposit date: | 2015-10-14 | | Release date: | 2015-11-11 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Analyses of Short-Chain Prenyltransferases Identify an Evolutionarily Conserved GFPPS Clade in Brassicaceae Plants.
Mol Plant, 9, 2016
|
|
8T0S
 
 | | Crystal structure of UBE2G2 adduct with phenethyl isothiocyanate (PEITC) at the Cys48 position | | Descriptor: | 1,2-ETHANEDIOL, E3 ubiquitin-protein ligase AMFR, Ubiquitin-conjugating enzyme E2 G2 | | Authors: | Wang, C, Shaw, G.X, Shi, G, Ji, X. | | Deposit date: | 2023-06-01 | | Release date: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structure of UBE2G2 adduct with phenethyl isothiocyanate (PEITC) at the Cys48 position
To be published
|
|
4M36
 
 | |
8WX8
 
 | | De novo design protein -T09 | | Descriptor: | De novo design protein -T09 | | Authors: | Wang, C, Wang, S, Liu, Y. | | Deposit date: | 2023-10-27 | | Release date: | 2024-10-09 | | Last modified: | 2025-05-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | De novo protein design with a denoising diffusion network independent of pretrained structure prediction models.
Nat.Methods, 21, 2024
|
|
1MHC
 
 | | MODEL OF MHC CLASS I H2-M3 WITH NONAPEPTIDE FROM RAT ND1 REFINED AT 2.3 ANGSTROMS RESOLUTION | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, MHC CLASS I ANTIGEN H2-M3, NONAPEPTIDE FROM RAT NADH DEHYDROGENASE | | Authors: | Wang, C.-R, Fischer Lindahl, K, Deisenhofer, J. | | Deposit date: | 1995-08-23 | | Release date: | 1996-01-29 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Nonclassical binding of formylated peptide in crystal structure of the MHC class Ib molecule H2-M3
Cell(Cambridge,Mass.), 82, 1995
|
|
4M38
 
 | |
4EKZ
 
 | | Crystal structure of reduced hPDI (abb'xa') | | Descriptor: | Protein disulfide-isomerase | | Authors: | Wang, C, Li, W, Ren, J, Ke, H, Gong, W, Feng, W, Wang, C.-C. | | Deposit date: | 2012-04-10 | | Release date: | 2013-04-10 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Structural insights into the redox-regulated dynamic conformations of human protein disulfide isomerase
Antioxid Redox Signal, 19, 2013
|
|
4EL1
 
 | | Crystal structure of oxidized hPDI (abb'xa') | | Descriptor: | Protein disulfide-isomerase | | Authors: | Wang, C, Li, W, Ren, J, Ke, H, Gong, W, Feng, W, Wang, C.-C. | | Deposit date: | 2012-04-10 | | Release date: | 2013-04-10 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.883 Å) | | Cite: | Structural insights into the redox-regulated dynamic conformations of human protein disulfide isomerase
Antioxid Redox Signal, 19, 2013
|
|
4M37
 
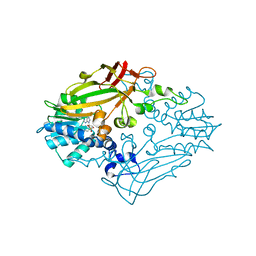 | |
7YDQ
 
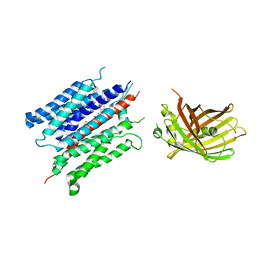 | | Structure of PfNT1(Y190A)-GFP in complex with GSK4 | | Descriptor: | 5-methyl-N-[2-(2-oxidanylideneazepan-1-yl)ethyl]-2-phenyl-1,3-oxazole-4-carboxamide, Nucleoside transporter 1,Green fluorescent protein | | Authors: | Wang, C, Yu, L.Y, Li, J.L, Ren, R.B, Deng, D. | | Deposit date: | 2022-07-04 | | Release date: | 2023-04-26 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (4.04 Å) | | Cite: | Structural basis of the substrate recognition and inhibition mechanism of Plasmodium falciparum nucleoside transporter PfENT1.
Nat Commun, 14, 2023
|
|
8DMF
 
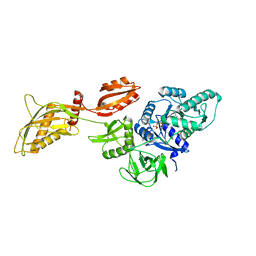 | | Cryo-EM structure of the ribosome-bound Bacteroides thetaiotaomicron EF-G2 | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Tetracycline resistance protein TetQ | | Authors: | Wang, C, Han, W, Groisman, E.A, Liu, J. | | Deposit date: | 2022-07-08 | | Release date: | 2023-01-04 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Gut colonization by Bacteroides requires translation by an EF-G paralog lacking GTPase activity.
Embo J., 2022
|
|
8EAN
 
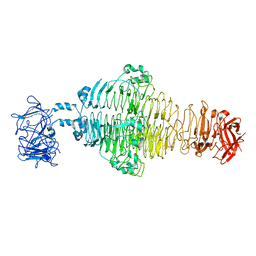 | |
8E4G
 
 | |
4H27
 
 | |
