7MJW
 
 | | Methylated MiaB in the complex with 5'-deoxyadenosine, methionine and RNA | | Descriptor: | 5'-DEOXYADENOSINE, FE3-S4 methylated cluster, IRON/SULFUR CLUSTER, ... | | Authors: | Esakova, O.A, Grove, T.L, Yennawar, N.H, Arcinas, A.J, Wang, B, Krebs, C, Almo, S.C, Booker, S.J. | | Deposit date: | 2021-04-20 | | Release date: | 2021-09-15 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural basis for tRNA methylthiolation by the radical SAM enzyme MiaB.
Nature, 597, 2021
|
|
7MJX
 
 | | MiaB in the complex with 5'-deoxyadenosine, methionine and RNA | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 5'-DEOXYADENOSINE, FE3-S4 CLUSTER, ... | | Authors: | Esakova, O.A, Grove, T.L, Yennawar, N.H, Arcinas, A.J, Wang, B, Krebs, C, Almo, S.C, Booker, S.J. | | Deposit date: | 2021-04-20 | | Release date: | 2021-09-15 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural basis for tRNA methylthiolation by the radical SAM enzyme MiaB.
Nature, 597, 2021
|
|
7MJY
 
 | | MiaB in the complex with s-adenosyl-L-homocysteine and RNA | | Descriptor: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, FE3-S4 CLUSTER, ... | | Authors: | Esakova, O.A, Grove, T.L, Yennawar, N.H, Arcinas, A.J, Wang, B, Krebs, C, Almo, S.C, Booker, S.J. | | Deposit date: | 2021-04-20 | | Release date: | 2021-09-15 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Structural basis for tRNA methylthiolation by the radical SAM enzyme MiaB.
Nature, 597, 2021
|
|
7MJZ
 
 | | The structure of MiaB with pentasulfide bridge | | Descriptor: | IRON/SULFUR CLUSTER, PENTASULFIDE-SULFUR, SODIUM ION, ... | | Authors: | Esakova, O.A, Grove, T.L, Yennawar, N.H, Arcinas, A.J, Wang, B, Krebs, C, Almo, S.C, Booker, S.J. | | Deposit date: | 2021-04-20 | | Release date: | 2021-09-15 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Structural basis for tRNA methylthiolation by the radical SAM enzyme MiaB.
Nature, 597, 2021
|
|
7M1S
 
 | |
8FDJ
 
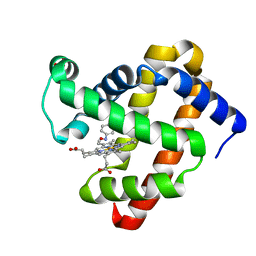 | | Wild-Type Sperm Whale Myoglobin in Complex with Nitrosobenzene | | Descriptor: | Myoglobin, NITROSOBENZENE, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Powell, S.M, Wang, B, Thomas, L.M, Richter-Addo, G.B. | | Deposit date: | 2022-12-03 | | Release date: | 2023-07-12 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structural investigations of heme protein derivatives resulting from reactions of aryl- and alkylhydroxylamines with human hemoglobin.
J.Inorg.Biochem., 246, 2023
|
|
8HB9
 
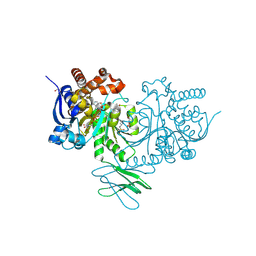 | | Crystal Structure of Human IDH1 R132H Mutant in Complex with NADPH and Compound IHMT-IDH1-053 | | Descriptor: | DI(HYDROXYETHYL)ETHER, GLYCEROL, Isocitrate dehydrogenase [NADP] cytoplasmic, ... | | Authors: | Guo, G, Wang, B, Liu, J, Liu, Q. | | Deposit date: | 2022-10-27 | | Release date: | 2023-05-24 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure-based discovery of IHMT-IDH1-053 as a potent irreversible IDH1 mutant selective inhibitor.
Eur.J.Med.Chem., 256, 2023
|
|
1A37
 
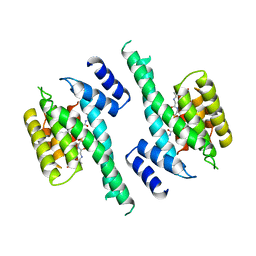 | | 14-3-3 PROTEIN ZETA BOUND TO PS-RAF259 PEPTIDE | | Descriptor: | 14-3-3 PROTEIN ZETA, PS-RAF259 PEPTIDE LSQRQRST(SEP)TPNVHM | | Authors: | Petosa, C, Masters, S.C, Pohl, J, Wang, B, Fu, H, Liddington, R.C. | | Deposit date: | 1998-01-28 | | Release date: | 1999-03-02 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | 14-3-3zeta binds a phosphorylated Raf peptide and an unphosphorylated peptide via its conserved amphipathic groove.
J.Biol.Chem., 273, 1998
|
|
1A38
 
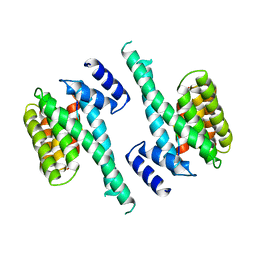 | | 14-3-3 PROTEIN ZETA BOUND TO R18 PEPTIDE | | Descriptor: | 14-3-3 PROTEIN ZETA, R18 PEPTIDE (PHCVPRDLSWLDLEANMCLP) | | Authors: | Petosa, C, Masters, S.C, Pohl, J, Wang, B, Fu, H, Liddington, R.C. | | Deposit date: | 1998-01-28 | | Release date: | 1999-03-02 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3.35 Å) | | Cite: | 14-3-3zeta binds a phosphorylated Raf peptide and an unphosphorylated peptide via its conserved amphipathic groove.
J.Biol.Chem., 273, 1998
|
|
8QMB
 
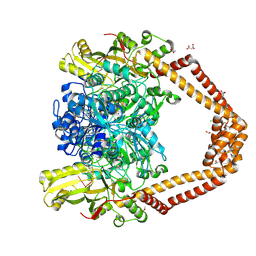 | | Nucleant-assisted 2.0 A resolution structure of the Streptococcus pneumoniae topoisomerase IV-V18mer DNA complex with the novel fluoroquinolone Delafloxacin | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, ACETATE ION, CHLORIDE ION, ... | | Authors: | Najmudin, S, Pan, X.S, Wang, B, Chayen, N.E, Fisher, L.M, Sanderson, M.R. | | Deposit date: | 2023-09-21 | | Release date: | 2024-10-02 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The nature of the molecular interactions at high resolution of the Streptococcus pneumoniae topoisomerase IV-DNA complex with the novel fluoroquinolone Delafloxacin.
To Be Published
|
|
8QMC
 
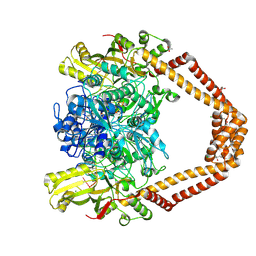 | | High resolution structure of the Streptococcus pneumoniae topoisomerase IV-complex with the V-site 18mer dsDNA and novel fluoroquinolone Delafloxacin | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, ACETATE ION, CHLORIDE ION, ... | | Authors: | Najmudin, S, Pan, X.S, Wang, B, Chayen, N.E, Fisher, L.M, Sanderson, M.R. | | Deposit date: | 2023-09-21 | | Release date: | 2024-10-02 | | Method: | X-RAY DIFFRACTION (2.402 Å) | | Cite: | The nature of the molecular interactions at high resolution of the Streptococcus pneumoniae topoisomerase IV-DNA complex with the novel fluoroquinolone Delafloxacin.
To Be Published
|
|
3HEI
 
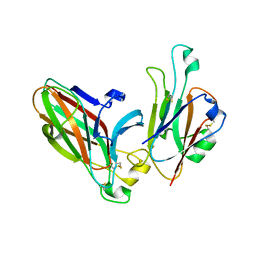 | | Ligand Recognition by A-Class Eph Receptors: Crystal Structures of the EphA2 Ligand-Binding Domain and the EphA2/ephrin-A1 Complex | | Descriptor: | Ephrin type-A receptor 2, Ephrin-A1 | | Authors: | Himanen, J.P, Goldgur, Y, Miao, H, Myshkin, E, Guo, H, Buck, M, Nguyen, M, Rajashankar, K.R, Wang, B, Nikolov, D.B. | | Deposit date: | 2009-05-08 | | Release date: | 2009-06-30 | | Last modified: | 2021-03-31 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Ligand recognition by A-class Eph receptors: crystal structures of the EphA2 ligand-binding domain and the EphA2/ephrin-A1 complex.
Embo Rep., 10, 2009
|
|
1O06
 
 | | Crystal structure of the Vps27p Ubiquitin Interacting Motif (UIM) | | Descriptor: | Vacuolar protein sorting-associated protein VPS27, ZINC ION | | Authors: | Fisher, R.D, Wang, B, Alam, S.L, Higginson, D.S, Rich, R, Myszka, D, Sundquist, W.I, Hill, C.P. | | Deposit date: | 2003-02-20 | | Release date: | 2003-07-22 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structure and ubiquitin binding of the ubiquitin-interacting motif.
J.Biol.Chem., 278, 2003
|
|
6VKJ
 
 | |
3HPN
 
 | | Ligand recognition by A-class EPH receptors: crystal structures of the EPHA2 ligand-binding domain and the EPHA2/EPHRIN-A1 complex | | Descriptor: | Ephrin type-A receptor 2 | | Authors: | Himanen, J.P, Goldgur, Y, Miao, H, Myshkin, E, Guo, H, Buck, M, Nguyen, M, Rajashankar, K.R, Wang, B, Nikolov, D.B. | | Deposit date: | 2009-06-04 | | Release date: | 2009-06-30 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.52 Å) | | Cite: | Ligand recognition by A-class Eph receptors: crystal structures of the EphA2 ligand-binding domain and the EphA2/ephrin-A1 complex.
Embo Rep., 10, 2009
|
|
5ANM
 
 | | Crystal structure of IgE Fc in complex with a neutralizing antibody | | Descriptor: | IG EPSILON CHAIN C REGION, IMMUNOGLOBULIN G, alpha-D-mannopyranose-(1-3)-alpha-D-mannopyranose-(1-6)-[alpha-D-mannopyranose-(1-3)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Cohen, E.S, Dobson, C.L, Kack, H, Wang, B, Sims, D.A, Lloyd, C.O, England, E, Rees, D.G, Guo, H, Karagiannis, S.N, O'Brien, S, Persdotter, S, Ekdahl, H, Butler, R, Keyes, F, Oakley, S, Carlsson, M, Briend, E, Wilkinson, T, Anderson, I.K, Monk, P.D, vonWachenfeldt, K, Eriksson, P.O, Gould, H.J, Vaughan, T.J, May, R.D. | | Deposit date: | 2015-09-07 | | Release date: | 2015-09-30 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | A Novel Ige-Neutralizing Antibody for the Treatment of Severe Uncontrolled Asthma.
Mabs, 6, 2015
|
|
7JSJ
 
 | | Structure of the NaCT-PF2 complex | | Descriptor: | (2R)-2-[2-(4-tert-butylphenyl)ethyl]-2-hydroxybutanedioic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, SODIUM ION, ... | | Authors: | Sauer, D.B, Wang, B, Song, J, Rice, W.J, Wang, D.N. | | Deposit date: | 2020-08-14 | | Release date: | 2021-02-24 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.12 Å) | | Cite: | Structure and inhibition mechanism of the human citrate transporter NaCT.
Nature, 591, 2021
|
|
7JSK
 
 | | Structure of the NaCT-Citrate complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CITRIC ACID, SODIUM ION, ... | | Authors: | Sauer, D.B, Wang, B, Song, J, Rice, W.J, Wang, D.N. | | Deposit date: | 2020-08-14 | | Release date: | 2021-02-24 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.04 Å) | | Cite: | Structure and inhibition mechanism of the human citrate transporter NaCT.
Nature, 591, 2021
|
|
7DCN
 
 | | Apo-citrate lyase phosphoribosyl-dephospho-CoA transferase | | Descriptor: | Probable apo-citrate lyase phosphoribosyl-dephospho-CoA transferase, SULFATE ION, ZINC ION | | Authors: | Xu, H, Wang, B, Su, X.D. | | Deposit date: | 2020-10-26 | | Release date: | 2021-11-03 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.695 Å) | | Cite: | Co-evolution-based prediction of metal-binding sites in proteomes by machine learning.
Nat.Chem.Biol., 19, 2023
|
|
7DCM
 
 | | Crystal structure of CITX | | Descriptor: | Probable apo-citrate lyase phosphoribosyl-dephospho-CoA transferase, ZINC ION | | Authors: | Xu, H, Wang, B, Su, X.D. | | Deposit date: | 2020-10-26 | | Release date: | 2021-11-03 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.495 Å) | | Cite: | Co-evolution-based prediction of metal-binding sites in proteomes by machine learning.
Nat.Chem.Biol., 19, 2023
|
|
6CF0
 
 | | Sperm Whale Myoglobin H64V Mutant with Nitrite | | Descriptor: | GLYCEROL, Myoglobin, NITRITE ION, ... | | Authors: | Powell, S.M, Wang, B, Thomas, L.M, Richter-Addo, G.B. | | Deposit date: | 2018-02-13 | | Release date: | 2018-05-02 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Nitrosyl Myoglobins and Their Nitrite Precursors: Crystal Structural and Quantum Mechanics and Molecular Mechanics Theoretical Investigations of Preferred Fe -NO Ligand Orientations in Myoglobin Distal Pockets.
Biochemistry, 57, 2018
|
|
7W06
 
 | | Itaconate inducible LysR-Type Transcriptional regulator (ITCR) in complex with itaconate (SeMet labeled), Space group C121. | | Descriptor: | 2-methylidenebutanedioic acid, CHLORIDE ION, SULFATE ION, ... | | Authors: | Sun, P.K, Wang, B, Wang, Z.X, Qi, S, Li, X.J. | | Deposit date: | 2021-11-17 | | Release date: | 2022-10-26 | | Last modified: | 2022-11-16 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | A genetically encoded fluorescent biosensor for detecting itaconate with subcellular resolution in living macrophages.
Nat Commun, 13, 2022
|
|
7TZL
 
 | | The DH dehydratase domain of AlnB | | Descriptor: | 3-oxoacyl-[acyl-carrier-protein] reductase | | Authors: | Swain, K, Blackson, W, Wang, B, Zhao, H, Nannenga, B.L. | | Deposit date: | 2022-02-16 | | Release date: | 2023-02-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | The programming of alpha,beta-polyene biosynthesis by a bacterial iterative type I polyketide synthase
To Be Published
|
|
7EK0
 
 | | Complex Structure of antibody BD-503 and RBD-N501Y of COVID-19 | | Descriptor: | Heavy Chain of BD-503, Light Chain of BD-503, Spike protein S1 | | Authors: | Xu, H, Wang, B, Zhao, T.N, Su, X.D. | | Deposit date: | 2021-04-03 | | Release date: | 2022-04-13 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure-based analyses of neutralization antibodies interacting with naturally occurring SARS-CoV-2 RBD variants.
Cell Res., 31, 2021
|
|
7EJY
 
 | | Complex Structure of antibody BD-503 and RBD of COVID-19 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy Chain of BD-503, Light Chain of BD-503, ... | | Authors: | Xu, H, Wang, B, Zhao, T.N, Su, X.D. | | Deposit date: | 2021-04-03 | | Release date: | 2022-04-13 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.04 Å) | | Cite: | Structure-based analyses of neutralization antibodies interacting with naturally occurring SARS-CoV-2 RBD variants.
Cell Res., 31, 2021
|
|
