3X3Z
 
 | | Copper amine oxidase from Arthrobacter globiformis: Aminoresorcinol form produced by anaerobic reduction with ethylamine hydrochloride | | Descriptor: | CHLORIDE ION, COPPER (II) ION, GLYCEROL, ... | | Authors: | Okajima, T, Nakanishi, S, Murakawa, T, Kataoka, M, Hayashi, H, Hamaguchi, A, Nakai, T, Kawano, Y, Yamaguchi, H, Tanizawa, K. | | Deposit date: | 2015-03-10 | | Release date: | 2015-08-19 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Probing the Catalytic Mechanism of Copper Amine Oxidase from Arthrobacter globiformis with Halide Ions.
J.Biol.Chem., 290, 2015
|
|
7COM
 
 | | Crystal structure of the SARS-CoV-2 main protease in complex with Boceprevir (space group P212121) | | Descriptor: | 3C-like proteinase, boceprevir (bound form) | | Authors: | Zeng, R, Qiao, J.X, Wang, Y.F, Li, Y.S, Yao, R, Liu, J.M, Zhou, Y.L, Chen, P, Yang, S.Y, Lei, J. | | Deposit date: | 2020-08-04 | | Release date: | 2020-08-19 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | SARS-CoV-2 M pro inhibitors with antiviral activity in a transgenic mouse model.
Science, 371, 2021
|
|
7D3I
 
 | | Crystal structure of SARS-CoV-2 main protease in complex with MI-23 | | Descriptor: | (3~{S},3~{a}~{S},6~{a}~{R})-2-[3-[3,5-bis(fluoranyl)phenyl]propanoyl]-~{N}-[(2~{S})-1-oxidanylidene-3-[(3~{S})-2-oxidanylidenepyrrolidin-3-yl]propan-2-yl]-3,3~{a},4,5,6,6~{a}-hexahydro-1~{H}-cyclopenta[c]pyrrole-3-carboxamide, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 3C-like proteinase | | Authors: | Zeng, R, Li, Y.S, Qiao, J.X, Wang, Y.F, Yang, S.Y, Lei, J. | | Deposit date: | 2020-09-19 | | Release date: | 2020-10-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.004 Å) | | Cite: | SARS-CoV-2 M pro inhibitors with antiviral activity in a transgenic mouse model.
Science, 371, 2021
|
|
7D4U
 
 | | ATP complex with double mutant cyclic trinucleotide synthase CdnD | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Cyclic AMP-AMP-GMP synthase | | Authors: | Yang, C.-S, Hou, M.-H, Tsai, C.-L, Wang, Y.-C, Ko, T.-P, Chen, Y. | | Deposit date: | 2020-09-24 | | Release date: | 2021-03-17 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure and functional implication of a bacterial cyclic AMP-AMP-GMP synthetase.
Nucleic Acids Res., 49, 2021
|
|
7D48
 
 | | apo-form cyclic trinucleotide synthase CdnD | | Descriptor: | Cyclic AMP-AMP-GMP synthase, SODIUM ION | | Authors: | Yang, C.-S, Hou, M.-H, Tsai, C.-L, Wang, Y.-C, Ko, T.-P, Chen, Y. | | Deposit date: | 2020-09-23 | | Release date: | 2021-03-17 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal structure and functional implication of a bacterial cyclic AMP-AMP-GMP synthetase.
Nucleic Acids Res., 49, 2021
|
|
7D4S
 
 | | apo-form cyclic trinucleotide synthase CdnD | | Descriptor: | Cyclic AMP-AMP-GMP synthase, DIPHOSPHOMETHYLPHOSPHONIC ACID ADENOSYL ESTER, MAGNESIUM ION | | Authors: | Yang, C.-S, Hou, M.-H, Tsai, C.-L, Wang, Y.-C, Ko, T.-P, Chen, Y. | | Deposit date: | 2020-09-24 | | Release date: | 2021-03-17 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Crystal structure and functional implication of a bacterial cyclic AMP-AMP-GMP synthetase.
Nucleic Acids Res., 49, 2021
|
|
7D4J
 
 | | ddATP complex of cyclic trinucleotide synthase CdnD | | Descriptor: | 2',3'-dideoxyadenosine triphosphate, Cyclic AMP-AMP-GMP synthase, MAGNESIUM ION | | Authors: | Yang, C.-S, Hou, M.-H, Tsai, C.-L, Wang, Y.-C, Ko, T.-P, Chen, Y. | | Deposit date: | 2020-09-24 | | Release date: | 2021-03-17 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Crystal structure and functional implication of a bacterial cyclic AMP-AMP-GMP synthetase.
Nucleic Acids Res., 49, 2021
|
|
3JRQ
 
 | | Crystal structure of (+)-ABA-bound PYL1 in complex with ABI1 | | Descriptor: | (2Z,4E)-5-[(1S)-1-hydroxy-2,6,6-trimethyl-4-oxocyclohex-2-en-1-yl]-3-methylpenta-2,4-dienoic acid, Protein phosphatase 2C 56, Putative uncharacterized protein At5g46790 | | Authors: | Miyazono, K, Miyakawa, T, Sawano, Y, Kubota, K, Tanokura, M. | | Deposit date: | 2009-09-08 | | Release date: | 2009-11-03 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis of abscisic acid signalling
Nature, 462, 2009
|
|
7D7E
 
 | | Structure of PKD1L3-CTD/PKD2L1 in apo state | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Polycystic kidney disease 2-like 1 protein, ... | | Authors: | Su, Q, Chen, M, Li, B, Wang, Y, Jing, D, Zhan, X, Yu, Y, Shi, Y. | | Deposit date: | 2020-10-03 | | Release date: | 2021-08-25 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural basis for Ca 2+ activation of the heteromeric PKD1L3/PKD2L1 channel.
Nat Commun, 12, 2021
|
|
7DBA
 
 | | RYX in complex with tubulin | | Descriptor: | 2-(1-methylindol-4-yl)-7-(3,4,5-trimethoxyphenyl)-1~{H}-benzimidazole, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, ... | | Authors: | Wu, C.Y, Wang, Y.X. | | Deposit date: | 2020-10-19 | | Release date: | 2021-10-20 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.461 Å) | | Cite: | RYX in complex with tubulin
To Be Published
|
|
7DBC
 
 | | PRA in complex with tubulin | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Wu, C.Y, Wang, Y.X. | | Deposit date: | 2020-10-19 | | Release date: | 2021-10-20 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | PRA in complex with tubulin
To Be Published
|
|
7DDC
 
 | | Crystal structure of SARS-CoV-2 main protease in complex with Tafenoquine | | Descriptor: | 3C-like proteinase, Tafenoquine | | Authors: | Chen, Y, Wang, Y.C, Yang, C.S, Hung, M.C. | | Deposit date: | 2020-10-28 | | Release date: | 2021-11-10 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.175 Å) | | Cite: | Tafenoquine and its derivatives as inhibitors for the severe acute respiratory syndrome coronavirus 2.
J.Biol.Chem., 298, 2022
|
|
3JRS
 
 | | Crystal structure of (+)-ABA-bound PYL1 | | Descriptor: | (2Z,4E)-5-[(1S)-1-hydroxy-2,6,6-trimethyl-4-oxocyclohex-2-en-1-yl]-3-methylpenta-2,4-dienoic acid, Putative uncharacterized protein At5g46790 | | Authors: | Miyazono, K, Miyakawa, T, Sawano, Y, Kubota, K, Tanokura, M. | | Deposit date: | 2009-09-08 | | Release date: | 2009-11-03 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural basis of abscisic acid signalling
Nature, 462, 2009
|
|
7EBL
 
 | | Bacterial STING in complex with c-di-GMP | | Descriptor: | 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), STING | | Authors: | Ko, T.-P, Wang, Y.-C, Yang, C.-S, Hou, M.-H, Chen, Y. | | Deposit date: | 2021-03-10 | | Release date: | 2021-09-15 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Crystal structure and functional implication of bacterial STING.
Nat Commun, 13, 2022
|
|
3D79
 
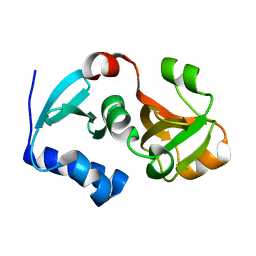 | | Crystal structure of hypothetical protein PH0734.1 from hyperthermophilic archaea Pyrococcus horikoshii OT3 | | Descriptor: | Putative uncharacterized protein PH0734 | | Authors: | Nishimura, Y, Miyazono, K, Sawano, Y, Makino, T, Nagata, K, Tanokura, M. | | Deposit date: | 2008-05-20 | | Release date: | 2008-12-09 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Crystal structure of hypothetical protein PH0734.1 from hyperthermophilic archaea Pyrococcus horikoshii OT3.
Proteins, 73, 2008
|
|
7F0A
 
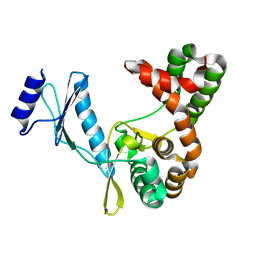 | |
7F0B
 
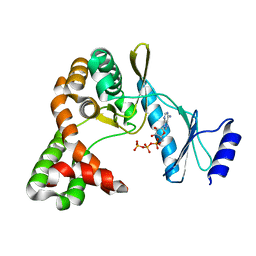 | | Crystal structure of capreomycin phosphotransferase in complex with ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Capreomycin phosphotransferase | | Authors: | Chang, C.Y, Pan, Y.C, Wang, Y.L, Toh, S.I. | | Deposit date: | 2021-06-03 | | Release date: | 2022-05-11 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Dual-Mechanism Confers Self-Resistance to the Antituberculosis Antibiotic Capreomycin.
Acs Chem.Biol., 17, 2022
|
|
7F0F
 
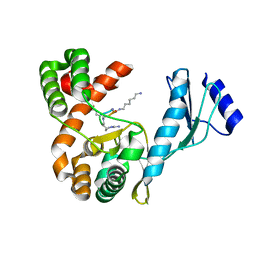 | | Crystal structure of capreomycin phosphotransferase in complex with CMN IIB | | Descriptor: | Capreomycin phosphotransferase, DPP-ALA-DPP-UAL-MYN-KBE | | Authors: | Chang, C.Y, Pan, Y.C, Wang, Y.L, Toh, S.I. | | Deposit date: | 2021-06-03 | | Release date: | 2022-05-11 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Dual-Mechanism Confers Self-Resistance to the Antituberculosis Antibiotic Capreomycin.
Acs Chem.Biol., 17, 2022
|
|
7F0C
 
 | | Crystal structure of capreomycin phosphotransferase in complex with CMN IIA | | Descriptor: | Capreomycin phosphotransferase, DPP-SER-DPP-UAL-MYN-KBE | | Authors: | Chang, C.Y, Pan, Y.C, Wang, Y.L, Toh, S.I. | | Deposit date: | 2021-06-03 | | Release date: | 2022-05-11 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Dual-Mechanism Confers Self-Resistance to the Antituberculosis Antibiotic Capreomycin.
Acs Chem.Biol., 17, 2022
|
|
3X40
 
 | | Copper amine oxidase from Arthrobacter globiformis: Product Schiff-base form produced by anaerobic reduction in the presence of sodium chloride | | Descriptor: | CHLORIDE ION, COPPER (II) ION, GLYCEROL, ... | | Authors: | Okajima, T, Nakanishi, S, Murakawa, T, Kataoka, M, Hayashi, H, Hamaguchi, A, Nakai, T, Kawano, Y, Yamaguchi, H, Tanizawa, K. | | Deposit date: | 2015-03-10 | | Release date: | 2015-08-19 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Probing the Catalytic Mechanism of Copper Amine Oxidase from Arthrobacter globiformis with Halide Ions.
J.Biol.Chem., 290, 2015
|
|
2DXB
 
 | | Recombinant thiocyanate hydrolase comprising partially-modified cobalt centers | | Descriptor: | COBALT (III) ION, PHOSPHATE ION, Thiocyanate hydrolase subunit alpha, ... | | Authors: | Arakawa, T, Kawano, Y, Katayama, Y, Yohda, M, Odaka, M. | | Deposit date: | 2006-08-25 | | Release date: | 2007-08-28 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural Basis for Catalytic Activation of Thiocyanate Hydrolase Involving Metal-Ligated Cysteine Modification
J.Am.Chem.Soc., 131, 2009
|
|
2DXC
 
 | | Recombinant thiocyanate hydrolase, fully-matured form | | Descriptor: | COBALT (III) ION, L(+)-TARTARIC ACID, Thiocyanate hydrolase subunit alpha, ... | | Authors: | Arakawa, T, Kawano, Y, Katayama, Y, Yohda, M, Odaka, M. | | Deposit date: | 2006-08-25 | | Release date: | 2007-08-28 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Basis for Catalytic Activation of Thiocyanate Hydrolase Involving Metal-Ligated Cysteine Modification
J.Am.Chem.Soc., 131, 2009
|
|
7C8B
 
 | | Crystal structure of the SARS-CoV-2 main protease in complex with Z-VAD(OMe)-FMK | | Descriptor: | 3C-like proteinase, CHLORIDE ION, Z-VAD(OMe)-FMK | | Authors: | Zeng, R, Qiao, J.X, Wang, Y.F, Li, Y.S, Yao, R, Yang, S.Y, Lei, J. | | Deposit date: | 2020-05-29 | | Release date: | 2020-09-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of the SARS-CoV-2 main protease in complex with Z-VAD(OMe)-FMK
To Be Published
|
|
7CX9
 
 | | Crystal structure of the SARS-CoV-2 main protease in complex with INZ-1 | | Descriptor: | 3-iodanyl-1~{H}-indazole-7-carbaldehyde, 3C-like proteinase, CHLORIDE ION, ... | | Authors: | Zeng, R, Liu, X.L, Qiao, J.X, Nan, J.S, Wang, Y.F, Li, Y.S, Yang, S.Y, Lei, J. | | Deposit date: | 2020-09-01 | | Release date: | 2020-09-16 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Crystal structure of the SARS-CoV-2 main protease in complex with INZ-1
To Be Published
|
|
7C3V
 
 | | Structure of a thermostable Alcohol dehydrogenase from Kluyveromyces polyspora(KpADH) | | Descriptor: | Alcohol dehydrogenase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Dai, W, Ni, Y, Xu, G, Liu, Y, Wang, Y, Zhou, J. | | Deposit date: | 2020-05-14 | | Release date: | 2021-05-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.20042944 Å) | | Cite: | Structure of a thermostable Alcohol dehydrogenase from Kluyveromyces polyspora(KpADH)
To Be Published
|
|
