3KHY
 
 | | Crystal Structure of a propionate kinase from Francisella tularensis subsp. tularensis SCHU S4 | | Descriptor: | Propionate kinase | | Authors: | Brunzelle, J.S, Skarina, T, Sharma, S, Wang, Y, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2009-10-31 | | Release date: | 2010-01-19 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.978 Å) | | Cite: | Crystal Structure of a propionate kinase from Francisella tularensis subsp. tularensis SCHU S4
To be Published
|
|
1U9M
 
 | | Crystal structure of F58W mutant of cytochrome b5 | | Descriptor: | Cytochrome b5, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Shan, L, Lu, J.-X, Gan, J.-H, Wang, Y.-H, Huang, Z.-X, Xia, Z.-X. | | Deposit date: | 2004-08-10 | | Release date: | 2005-02-01 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of the F58W mutant of cytochrome b5: the mutation leads to multiple conformations and weakens stacking interactions.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
5GQ9
 
 | |
3OU4
 
 | | MDR769 HIV-1 protease complexed with TF/PR hepta-peptide | | Descriptor: | HIV-1 protease, TF/PR substrate peptide | | Authors: | Liu, Z, Wang, Y, Brunzelle, J, Kovari, I.A, Kovari, L.C. | | Deposit date: | 2010-09-14 | | Release date: | 2011-03-30 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Nine Crystal Structures Determine the Substrate Envelope of the MDR HIV-1 Protease.
Protein J., 30, 2011
|
|
3KN0
 
 | | Structure of BACE bound to SCH708236 | | Descriptor: | 3-[2-(3-{[(furan-2-ylmethyl)(methyl)amino]methyl}phenyl)ethyl]pyridin-2-amine, Beta-secretase 1, L(+)-TARTARIC ACID | | Authors: | Strickland, C, Wang, Y. | | Deposit date: | 2009-11-11 | | Release date: | 2010-01-19 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Application of Fragment-Based NMR Screening, X-ray Crystallography, Structure-Based Design, and Focused Chemical Library Design to Identify Novel muM Leads for the Development of nM BACE-1 (beta-Site APP Cleaving Enzyme 1) Inhibitors.
J.Med.Chem., 53, 2010
|
|
3KMX
 
 | | Structure of BACE bound to SCH346572 | | Descriptor: | 4-butoxy-3-chlorobenzyl imidothiocarbamate, Beta-secretase 1 | | Authors: | Strickland, C, Wang, Y. | | Deposit date: | 2009-11-11 | | Release date: | 2010-01-19 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Application of Fragment-Based NMR Screening, X-ray Crystallography, Structure-Based Design, and Focused Chemical Library Design to Identify Novel muM Leads for the Development of nM BACE-1 (beta-Site APP Cleaving Enzyme 1) Inhibitors.
J.Med.Chem., 53, 2010
|
|
3OU3
 
 | | MDR769 HIV-1 protease complexed with PR/RT hepta-peptide | | Descriptor: | HIV-1 protease, PR/RT substrate peptide | | Authors: | Liu, Z, Wang, Y, Brunzelle, J, Kovari, I.A, Kovari, L.C. | | Deposit date: | 2010-09-14 | | Release date: | 2011-03-30 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Nine Crystal Structures Determine the Substrate Envelope of the MDR HIV-1 Protease.
Protein J., 30, 2011
|
|
3OUC
 
 | | MDR769 HIV-1 protease complexed with p2/NC hepta-peptide | | Descriptor: | MDR HIV-1 protease, p2/NC substrate peptide | | Authors: | Liu, Z, Wang, Y, Brunzelle, J, Kovari, I.A, Kovari, L.C. | | Deposit date: | 2010-09-14 | | Release date: | 2011-03-30 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Nine Crystal Structures Determine the Substrate Envelope of the MDR HIV-1 Protease.
Protein J., 30, 2011
|
|
3OY3
 
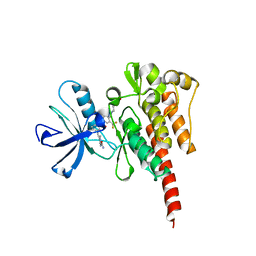 | | Crystal structure of ABL T315I mutant kinase domain bound with a DFG-out inhibitor AP24589 | | Descriptor: | 5-[(5-{[4-{[4-(2-hydroxyethyl)piperazin-1-yl]methyl}-3-(trifluoromethyl)phenyl]carbamoyl}-2-methylphenyl)ethynyl]-1-methyl-1H-imidazole-2-carboxamide, Tyrosine-protein kinase ABL1 | | Authors: | Zhou, T, Commodore, L, Huang, W.S, Wang, Y, Thomas, M, Keats, J, Xu, Q, Rivera, V, Shakespeare, W.C, Clackson, T, Dalgarno, D.C, Zhu, X. | | Deposit date: | 2010-09-22 | | Release date: | 2010-12-15 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural Mechanism of the Pan-BCR-ABL Inhibitor Ponatinib (AP24534): Lessons for Overcoming Kinase Inhibitor Resistance.
Chem.Biol.Drug Des., 77, 2011
|
|
1U9U
 
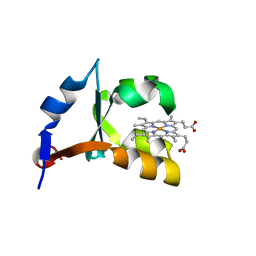 | | Crystal structure of F58Y mutant of cytochrome b5 | | Descriptor: | Cytochrome b5, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Shan, L, Lu, J.-X, Gan, J.-H, Wang, Y.-H, Huang, Z.-X, Xia, Z.-X. | | Deposit date: | 2004-08-11 | | Release date: | 2005-02-01 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Structure of the F58W mutant of cytochrome b5: the mutation leads to multiple conformations and weakens stacking interactions.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
3OUB
 
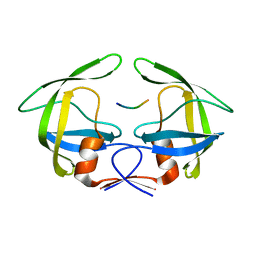 | | MDR769 HIV-1 protease complexed with NC/p1 hepta-peptide | | Descriptor: | MDR HIV-1 protease, NC/p1 substrate peptide | | Authors: | Liu, Z, Wang, Y, Brunzelle, J, Kovari, I.A, Kovari, L.C. | | Deposit date: | 2010-09-14 | | Release date: | 2011-03-30 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Nine Crystal Structures Determine the Substrate Envelope of the MDR HIV-1 Protease.
Protein J., 30, 2011
|
|
3P8P
 
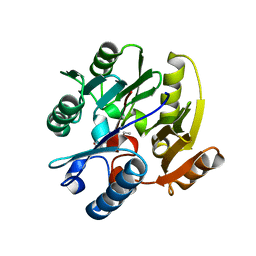 | | Crystal Structure of Human Dimethylarginine Dimethylaminohydrolase-1 (DDAH-1) variant C274S bound with N5-(1-iminopentyl)-L-ornithine | | Descriptor: | N(G),N(G)-dimethylarginine dimethylaminohydrolase 1, N~5~-[(1E)-pentanimidoyl]-L-ornithine | | Authors: | Monzingo, A.F, Lluis, M, Wang, Y, Fast, W, Robertus, J.D. | | Deposit date: | 2010-10-14 | | Release date: | 2010-11-10 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Characterization of C-Alkyl Amidines as Bioavailable Covalent Reversible Inhibitors of Human DDAH-1.
Chemmedchem, 6, 2011
|
|
3P8E
 
 | | Crystal structure of human DIMETHYLARGININE DIMETHYLAMINOHYDROLASE-1 (DDAH-1) covalently bound with N5-(1-iminopentyl)-L-ornithine | | Descriptor: | N(G),N(G)-dimethylarginine dimethylaminohydrolase 1, N~5~-[(1S)-1-aminopentyl]-L-ornithine | | Authors: | Lluis, M, Wang, Y, Monzingo, A.F, Fast, W, Robertus, J.D. | | Deposit date: | 2010-10-13 | | Release date: | 2010-11-10 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.4946 Å) | | Cite: | Characterization of C-Alkyl Amidines as Bioavailable Covalent Reversible Inhibitors of Human DDAH-1.
Chemmedchem, 6, 2011
|
|
3OUD
 
 | | MDR769 HIV-1 protease complexed with CA/p2 hepta-peptide | | Descriptor: | CA/p2 substrate peptide, MDR HIV-1 protease | | Authors: | Liu, Z, Wang, Y, Brunzelle, J, Kovari, I.A, Kovari, L.C. | | Deposit date: | 2010-09-14 | | Release date: | 2011-03-30 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Nine Crystal Structures Determine the Substrate Envelope of the MDR HIV-1 Protease.
Protein J., 30, 2011
|
|
5YEF
 
 | | Crystal structure of CTCF ZFs2-8-Hs5-1aE | | Descriptor: | DNA (27-MER), Transcriptional repressor CTCF, ZINC ION | | Authors: | Yin, M, Wang, J, Wang, M, Li, X, Wang, Y. | | Deposit date: | 2017-09-17 | | Release date: | 2017-11-29 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.807 Å) | | Cite: | Molecular mechanism of directional CTCF recognition of a diverse range of genomic sites
Cell Res., 27, 2017
|
|
7TRD
 
 | | Human telomerase catalytic core structure at 3.3 Angstrom | | Descriptor: | Telomerase RNA, partial sequence, Telomerase reverse transcriptase, ... | | Authors: | Liu, B, He, Y, Wang, Y, Song, H, Zhou, Z.H, Feigon, J. | | Deposit date: | 2022-01-28 | | Release date: | 2022-04-20 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structure of active human telomerase with telomere shelterin protein TPP1.
Nature, 604, 2022
|
|
7TRE
 
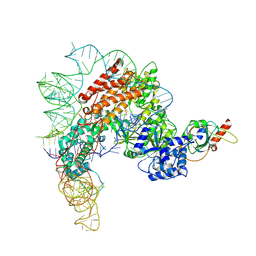 | | Human telomerase catalytic core with shelterin protein TPP1 | | Descriptor: | Adrenocortical dysplasia protein homolog, Telomerase RNA, partial sequence, ... | | Authors: | Liu, B, He, Y, Wang, Y, Song, H, Zhou, Z.H, Feigon, J. | | Deposit date: | 2022-01-28 | | Release date: | 2022-04-20 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structure of active human telomerase with telomere shelterin protein TPP1.
Nature, 604, 2022
|
|
7TRF
 
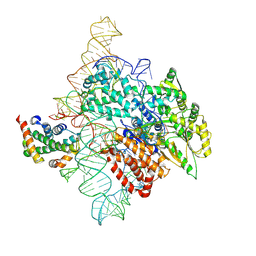 | | Human telomerase catalytic core RNP with H2A/H2B | | Descriptor: | Histone H2A, Histone H2B type 1-C/E/F/G/I, Telomerase RNA, ... | | Authors: | Liu, B, He, Y, Wang, Y, Song, H, Zhou, Z.H, Feigon, J. | | Deposit date: | 2022-01-28 | | Release date: | 2022-04-20 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structure of active human telomerase with telomere shelterin protein TPP1.
Nature, 604, 2022
|
|
7TRC
 
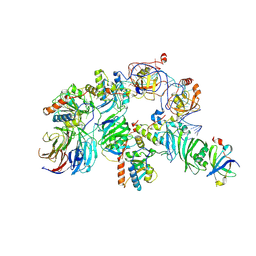 | | Human telomerase H/ACA RNP at 3.3 Angstrom | | Descriptor: | H/ACA ribonucleoprotein complex subunit 1, H/ACA ribonucleoprotein complex subunit 2, H/ACA ribonucleoprotein complex subunit 3, ... | | Authors: | Liu, B, He, Y, Wang, Y, Song, H, Zhou, Z.H, Feigon, J. | | Deposit date: | 2022-01-28 | | Release date: | 2022-04-20 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structure of active human telomerase with telomere shelterin protein TPP1.
Nature, 604, 2022
|
|
8KDE
 
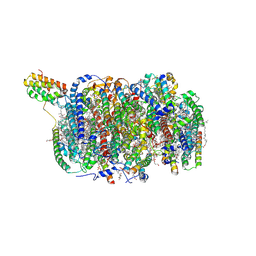 | | Cryo-EM structure of an intermediate-state complex during the process of photosystem II repair | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Li, A, Wang, Y, Liu, Z. | | Deposit date: | 2023-08-09 | | Release date: | 2024-06-19 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Structural basis for an early stage of the photosystem II repair cycle in Chlamydomonas reinhardtii.
Nat Commun, 15, 2024
|
|
5VEX
 
 | | Structure of the human GLP-1 receptor complex with NNC0640 | | Descriptor: | 4-{[(4-cyclohexylphenyl){[3-(methylsulfonyl)phenyl]carbamoyl}amino]methyl}-N-(1H-tetrazol-5-yl)benzamide, Glucagon-like peptide 1 receptor, Endolysin chimera | | Authors: | Song, G, Yang, D, Wang, Y, Graaf, C.D, Zhou, Q, Jiang, S, Liu, K, Cai, X, Dai, A, Lin, G, Liu, D, Wu, F, Wu, Y, Zhao, S, Ye, L, Han, G.W, Lau, J, Wu, B, Hanson, M.A, Liu, Z.-J, Wang, M.-W, Stevens, R.C. | | Deposit date: | 2017-04-05 | | Release date: | 2017-05-17 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Human GLP-1 receptor transmembrane domain structure in complex with allosteric modulators.
Nature, 546, 2017
|
|
8WRB
 
 | | Lysophosphatidylserine receptor GPR34-Gi complex | | Descriptor: | Antibody fragment scFv16, CHOLESTEROL, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Gong, W, Liu, G, Li, X, Wang, Y, Zhang, X. | | Deposit date: | 2023-10-13 | | Release date: | 2023-11-08 | | Last modified: | 2023-12-20 | | Method: | ELECTRON MICROSCOPY (2.91 Å) | | Cite: | Structural basis for ligand recognition and signaling of the lysophosphatidylserine receptors GPR34 and GPR174.
Plos Biol., 21, 2023
|
|
6JEA
 
 | | crystal structure of a beta-N-acetylhexosaminidase | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-N-acetylhexosaminidase, ZINC ION | | Authors: | Chen, X, Wang, J.C, Liu, M.J, Yang, W.Y, Wang, Y.Z, Tang, R.P, Zhang, M. | | Deposit date: | 2019-02-04 | | Release date: | 2019-03-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.275 Å) | | Cite: | Crystallographic evidence for substrate-assisted catalysis of beta-N-acetylhexosaminidas from Akkermansia muciniphila.
Biochem. Biophys. Res. Commun., 511, 2019
|
|
8SNL
 
 | | Structure of human ADAM17/iRhom2 sheddase complex | | Descriptor: | CALCIUM ION, Disintegrin and metalloproteinase domain-containing protein 17, Inactive rhomboid protein 2, ... | | Authors: | Zhao, H, Dai, Y, Wang, Y, Lee, C.H. | | Deposit date: | 2023-04-27 | | Release date: | 2024-05-29 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (2.78 Å) | | Cite: | Structure of human ADAM17/iRhom2 sheddase complex
To Be Published
|
|
8SNN
 
 | | Structure of mature human ADAM17/iRhom2 sheddase complex, conformation 1 | | Descriptor: | CALCIUM ION, Disintegrin and metalloproteinase domain-containing protein 17, Inactive rhomboid protein 2 | | Authors: | Zhao, H, Dai, Y, Wang, Y, Lee, C.H. | | Deposit date: | 2023-04-27 | | Release date: | 2024-05-29 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.32 Å) | | Cite: | Structure of mature human ADAM17/iRhom2 sheddase complex, conformation 1
To Be Published
|
|
